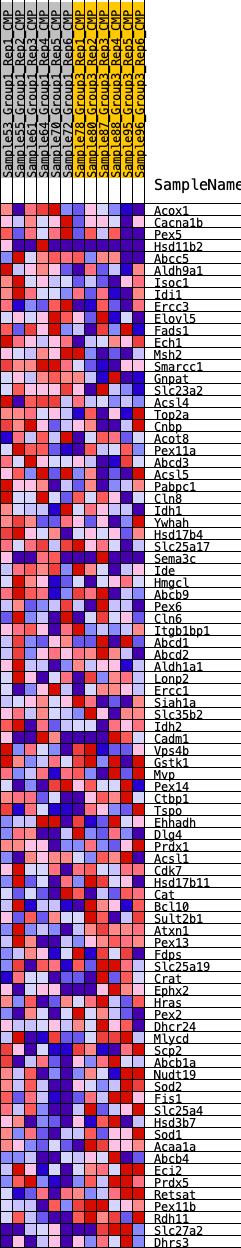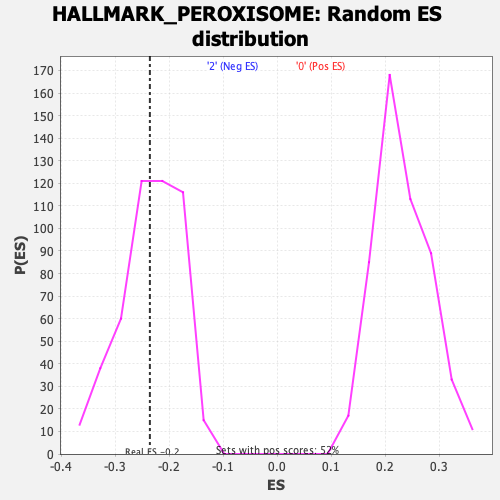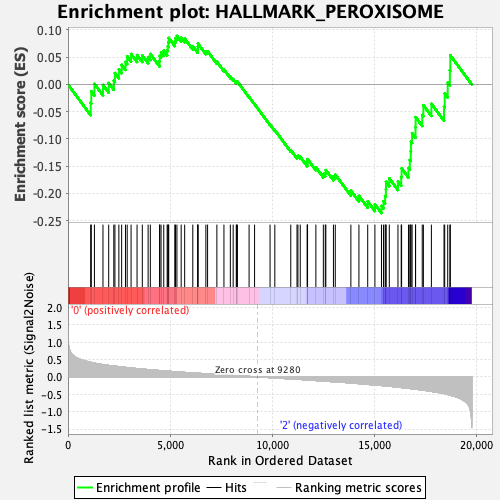
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.23609471 |
| Normalized Enrichment Score (NES) | -1.0080621 |
| Nominal p-value | 0.45661157 |
| FDR q-value | 0.81505156 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Acox1 | 1111 | 0.421 | -0.0337 | No |
| 2 | Cacna1b | 1140 | 0.416 | -0.0126 | No |
| 3 | Pex5 | 1294 | 0.398 | 0.0011 | No |
| 4 | Hsd11b2 | 1712 | 0.355 | -0.0009 | No |
| 5 | Abcc5 | 1993 | 0.330 | 0.0028 | No |
| 6 | Aldh9a1 | 2242 | 0.313 | 0.0071 | No |
| 7 | Isoc1 | 2297 | 0.309 | 0.0211 | No |
| 8 | Idi1 | 2490 | 0.297 | 0.0274 | No |
| 9 | Ercc3 | 2627 | 0.288 | 0.0360 | No |
| 10 | Elovl5 | 2814 | 0.274 | 0.0414 | No |
| 11 | Fads1 | 2898 | 0.269 | 0.0517 | No |
| 12 | Ech1 | 3088 | 0.257 | 0.0560 | No |
| 13 | Msh2 | 3381 | 0.240 | 0.0542 | No |
| 14 | Smarcc1 | 3638 | 0.225 | 0.0533 | No |
| 15 | Gnpat | 3923 | 0.211 | 0.0503 | No |
| 16 | Slc23a2 | 4031 | 0.205 | 0.0560 | No |
| 17 | Acsl4 | 4484 | 0.183 | 0.0429 | No |
| 18 | Top2a | 4492 | 0.183 | 0.0525 | No |
| 19 | Cnbp | 4556 | 0.180 | 0.0590 | No |
| 20 | Acot8 | 4688 | 0.176 | 0.0619 | No |
| 21 | Pex11a | 4855 | 0.167 | 0.0625 | No |
| 22 | Abcd3 | 4885 | 0.166 | 0.0700 | No |
| 23 | Acsl5 | 4915 | 0.165 | 0.0774 | No |
| 24 | Pabpc1 | 4932 | 0.164 | 0.0854 | No |
| 25 | Cln8 | 5229 | 0.150 | 0.0785 | No |
| 26 | Idh1 | 5264 | 0.149 | 0.0848 | No |
| 27 | Ywhah | 5333 | 0.145 | 0.0892 | No |
| 28 | Hsd17b4 | 5541 | 0.135 | 0.0860 | No |
| 29 | Slc25a17 | 5712 | 0.129 | 0.0843 | No |
| 30 | Sema3c | 6110 | 0.115 | 0.0704 | No |
| 31 | Ide | 6351 | 0.108 | 0.0640 | No |
| 32 | Hmgcl | 6357 | 0.107 | 0.0695 | No |
| 33 | Abcb9 | 6364 | 0.106 | 0.0750 | No |
| 34 | Pex6 | 6750 | 0.092 | 0.0604 | No |
| 35 | Cln6 | 6834 | 0.089 | 0.0610 | No |
| 36 | Itgb1bp1 | 7289 | 0.072 | 0.0418 | No |
| 37 | Abcd1 | 7635 | 0.059 | 0.0274 | No |
| 38 | Abcd2 | 7948 | 0.049 | 0.0142 | No |
| 39 | Aldh1a1 | 8087 | 0.044 | 0.0096 | No |
| 40 | Lonp2 | 8239 | 0.038 | 0.0040 | No |
| 41 | Ercc1 | 8256 | 0.038 | 0.0052 | No |
| 42 | Siah1a | 8295 | 0.037 | 0.0053 | No |
| 43 | Slc35b2 | 8866 | 0.015 | -0.0229 | No |
| 44 | Idh2 | 9135 | 0.006 | -0.0362 | No |
| 45 | Cadm1 | 9893 | -0.020 | -0.0735 | No |
| 46 | Vps4b | 10126 | -0.029 | -0.0838 | No |
| 47 | Gstk1 | 10907 | -0.056 | -0.1203 | No |
| 48 | Mvp | 11212 | -0.065 | -0.1323 | No |
| 49 | Pex14 | 11257 | -0.067 | -0.1309 | No |
| 50 | Ctbp1 | 11371 | -0.071 | -0.1327 | No |
| 51 | Tspo | 11714 | -0.085 | -0.1455 | No |
| 52 | Ehhadh | 11718 | -0.085 | -0.1411 | No |
| 53 | Dlg4 | 11729 | -0.086 | -0.1369 | No |
| 54 | Prdx1 | 12136 | -0.103 | -0.1520 | No |
| 55 | Acsl1 | 12502 | -0.118 | -0.1641 | No |
| 56 | Cdk7 | 12596 | -0.122 | -0.1623 | No |
| 57 | Hsd17b11 | 12624 | -0.123 | -0.1570 | No |
| 58 | Cat | 12996 | -0.138 | -0.1684 | No |
| 59 | Bcl10 | 13086 | -0.139 | -0.1654 | No |
| 60 | Sult2b1 | 13850 | -0.173 | -0.1948 | No |
| 61 | Atxn1 | 14244 | -0.193 | -0.2043 | No |
| 62 | Pex13 | 14674 | -0.215 | -0.2145 | No |
| 63 | Fdps | 15026 | -0.232 | -0.2198 | No |
| 64 | Slc25a19 | 15348 | -0.246 | -0.2228 | Yes |
| 65 | Crat | 15446 | -0.252 | -0.2141 | Yes |
| 66 | Ephx2 | 15527 | -0.256 | -0.2044 | Yes |
| 67 | Hras | 15564 | -0.258 | -0.1923 | Yes |
| 68 | Pex2 | 15569 | -0.258 | -0.1785 | Yes |
| 69 | Dhcr24 | 15731 | -0.266 | -0.1723 | Yes |
| 70 | Mlycd | 16155 | -0.294 | -0.1779 | Yes |
| 71 | Scp2 | 16314 | -0.304 | -0.1695 | Yes |
| 72 | Abcb1a | 16334 | -0.306 | -0.1539 | Yes |
| 73 | Nudt19 | 16670 | -0.328 | -0.1532 | Yes |
| 74 | Sod2 | 16735 | -0.332 | -0.1385 | Yes |
| 75 | Fis1 | 16782 | -0.335 | -0.1227 | Yes |
| 76 | Slc25a4 | 16790 | -0.336 | -0.1049 | Yes |
| 77 | Hsd3b7 | 16847 | -0.340 | -0.0893 | Yes |
| 78 | Sod1 | 17014 | -0.352 | -0.0787 | Yes |
| 79 | Acaa1a | 17019 | -0.353 | -0.0599 | Yes |
| 80 | Abcb4 | 17344 | -0.378 | -0.0559 | Yes |
| 81 | Eci2 | 17400 | -0.383 | -0.0380 | Yes |
| 82 | Prdx5 | 17792 | -0.417 | -0.0353 | Yes |
| 83 | Retsat | 18410 | -0.477 | -0.0408 | Yes |
| 84 | Pex11b | 18444 | -0.481 | -0.0165 | Yes |
| 85 | Rdh11 | 18591 | -0.505 | 0.0034 | Yes |
| 86 | Slc27a2 | 18697 | -0.520 | 0.0262 | Yes |
| 87 | Dhrs3 | 18719 | -0.524 | 0.0534 | Yes |