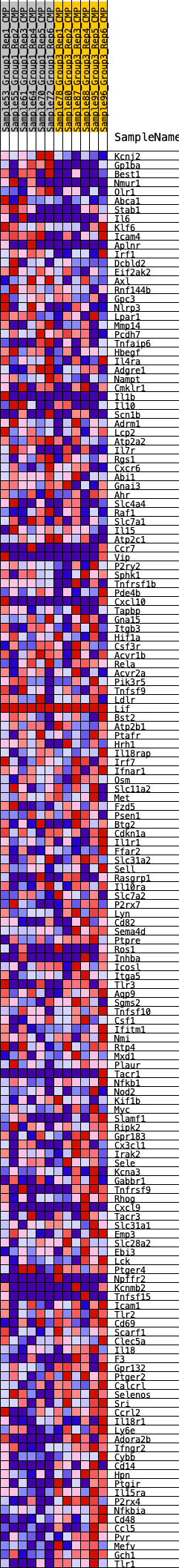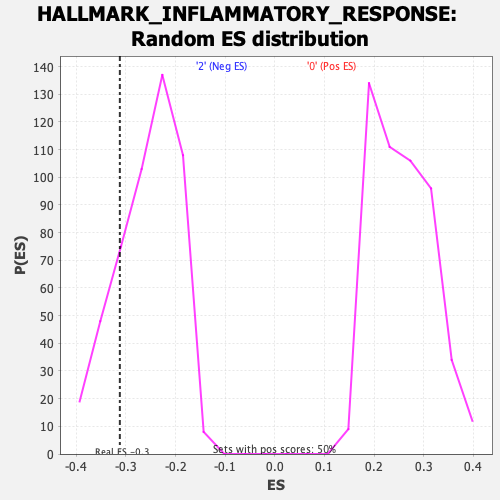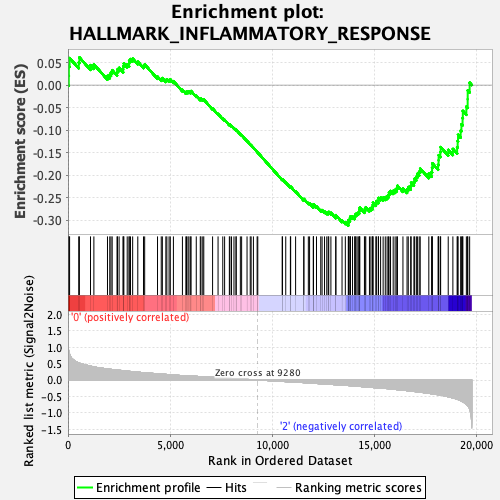
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.31208393 |
| Normalized Enrichment Score (NES) | -1.2256224 |
| Nominal p-value | 0.1746988 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.874 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Kcnj2 | 38 | 0.895 | 0.0204 | No |
| 2 | Gp1ba | 42 | 0.872 | 0.0421 | No |
| 3 | Best1 | 78 | 0.786 | 0.0600 | No |
| 4 | Nmur1 | 526 | 0.521 | 0.0502 | No |
| 5 | Olr1 | 557 | 0.512 | 0.0615 | No |
| 6 | Abca1 | 1102 | 0.422 | 0.0443 | No |
| 7 | Stab1 | 1267 | 0.401 | 0.0459 | No |
| 8 | Il6 | 1933 | 0.335 | 0.0204 | No |
| 9 | Klf6 | 2045 | 0.327 | 0.0229 | No |
| 10 | Icam4 | 2103 | 0.323 | 0.0281 | No |
| 11 | Aplnr | 2163 | 0.318 | 0.0330 | No |
| 12 | Irf1 | 2401 | 0.302 | 0.0285 | No |
| 13 | Dcbld2 | 2423 | 0.301 | 0.0350 | No |
| 14 | Eif2ak2 | 2502 | 0.296 | 0.0384 | No |
| 15 | Axl | 2700 | 0.282 | 0.0354 | No |
| 16 | Rnf144b | 2704 | 0.281 | 0.0423 | No |
| 17 | Gpc3 | 2735 | 0.279 | 0.0477 | No |
| 18 | Nlrp3 | 2899 | 0.269 | 0.0461 | No |
| 19 | Lpar1 | 2992 | 0.263 | 0.0480 | No |
| 20 | Mmp14 | 2995 | 0.263 | 0.0545 | No |
| 21 | Pcdh7 | 3060 | 0.260 | 0.0577 | No |
| 22 | Tnfaip6 | 3166 | 0.253 | 0.0587 | No |
| 23 | Hbegf | 3417 | 0.238 | 0.0519 | No |
| 24 | Il4ra | 3696 | 0.223 | 0.0433 | No |
| 25 | Adgre1 | 3754 | 0.220 | 0.0459 | No |
| 26 | Nampt | 4383 | 0.189 | 0.0186 | No |
| 27 | Cmklr1 | 4565 | 0.180 | 0.0139 | No |
| 28 | Il1b | 4620 | 0.178 | 0.0156 | No |
| 29 | Il10 | 4791 | 0.171 | 0.0112 | No |
| 30 | Scn1b | 4840 | 0.168 | 0.0129 | No |
| 31 | Adrm1 | 4949 | 0.163 | 0.0115 | No |
| 32 | Lcp2 | 5009 | 0.160 | 0.0125 | No |
| 33 | Atp2a2 | 5161 | 0.153 | 0.0086 | No |
| 34 | Il7r | 5602 | 0.134 | -0.0105 | No |
| 35 | Rgs1 | 5771 | 0.127 | -0.0159 | No |
| 36 | Cxcr6 | 5802 | 0.126 | -0.0143 | No |
| 37 | Abi1 | 5874 | 0.124 | -0.0148 | No |
| 38 | Gnai3 | 5924 | 0.121 | -0.0143 | No |
| 39 | Ahr | 6007 | 0.118 | -0.0155 | No |
| 40 | Slc4a4 | 6022 | 0.118 | -0.0132 | No |
| 41 | Raf1 | 6280 | 0.111 | -0.0236 | No |
| 42 | Slc7a1 | 6476 | 0.102 | -0.0310 | No |
| 43 | Il15 | 6510 | 0.101 | -0.0301 | No |
| 44 | Atp2c1 | 6599 | 0.097 | -0.0322 | No |
| 45 | Ccr7 | 6648 | 0.096 | -0.0322 | No |
| 46 | Vip | 7079 | 0.080 | -0.0522 | No |
| 47 | P2ry2 | 7347 | 0.070 | -0.0640 | No |
| 48 | Sphk1 | 7578 | 0.061 | -0.0742 | No |
| 49 | Tnfrsf1b | 7668 | 0.058 | -0.0773 | No |
| 50 | Pde4b | 7900 | 0.050 | -0.0879 | No |
| 51 | Cxcl10 | 7927 | 0.050 | -0.0879 | No |
| 52 | Tapbp | 8006 | 0.047 | -0.0908 | No |
| 53 | Gna15 | 8126 | 0.042 | -0.0958 | No |
| 54 | Itgb3 | 8227 | 0.038 | -0.0999 | No |
| 55 | Hif1a | 8234 | 0.038 | -0.0992 | No |
| 56 | Csf3r | 8438 | 0.031 | -0.1088 | No |
| 57 | Acvr1b | 8501 | 0.029 | -0.1113 | No |
| 58 | Rela | 8766 | 0.019 | -0.1242 | No |
| 59 | Acvr2a | 8922 | 0.013 | -0.1318 | No |
| 60 | Pik3r5 | 8966 | 0.011 | -0.1337 | No |
| 61 | Tnfsf9 | 9078 | 0.008 | -0.1392 | No |
| 62 | Ldlr | 9260 | 0.001 | -0.1484 | No |
| 63 | Lif | 9292 | 0.000 | -0.1500 | No |
| 64 | Bst2 | 10486 | -0.042 | -0.2097 | No |
| 65 | Atp2b1 | 10503 | -0.043 | -0.2095 | No |
| 66 | Ptafr | 10661 | -0.048 | -0.2163 | No |
| 67 | Hrh1 | 10888 | -0.056 | -0.2264 | No |
| 68 | Il18rap | 10902 | -0.056 | -0.2257 | No |
| 69 | Irf7 | 11144 | -0.063 | -0.2364 | No |
| 70 | Ifnar1 | 11546 | -0.078 | -0.2549 | No |
| 71 | Osm | 11547 | -0.079 | -0.2529 | No |
| 72 | Slc11a2 | 11767 | -0.087 | -0.2619 | No |
| 73 | Met | 11828 | -0.090 | -0.2627 | No |
| 74 | Fzd5 | 12014 | -0.099 | -0.2697 | No |
| 75 | Psen1 | 12016 | -0.099 | -0.2672 | No |
| 76 | Btg2 | 12017 | -0.099 | -0.2648 | No |
| 77 | Cdkn1a | 12166 | -0.105 | -0.2697 | No |
| 78 | Il1r1 | 12375 | -0.114 | -0.2775 | No |
| 79 | Ffar2 | 12440 | -0.116 | -0.2778 | No |
| 80 | Slc31a2 | 12560 | -0.120 | -0.2809 | No |
| 81 | Sell | 12670 | -0.125 | -0.2833 | No |
| 82 | Rasgrp1 | 12740 | -0.127 | -0.2836 | No |
| 83 | Il10ra | 12766 | -0.129 | -0.2817 | No |
| 84 | Slc7a2 | 12872 | -0.133 | -0.2837 | No |
| 85 | P2rx7 | 13110 | -0.141 | -0.2923 | No |
| 86 | Lyn | 13115 | -0.141 | -0.2890 | No |
| 87 | Cd82 | 13417 | -0.154 | -0.3005 | No |
| 88 | Sema4d | 13578 | -0.161 | -0.3046 | No |
| 89 | Ptpre | 13726 | -0.168 | -0.3079 | Yes |
| 90 | Ros1 | 13730 | -0.168 | -0.3038 | Yes |
| 91 | Inhba | 13733 | -0.168 | -0.2997 | Yes |
| 92 | Icosl | 13803 | -0.171 | -0.2990 | Yes |
| 93 | Itga5 | 13811 | -0.172 | -0.2950 | Yes |
| 94 | Tlr3 | 13829 | -0.172 | -0.2916 | Yes |
| 95 | Aqp9 | 13931 | -0.177 | -0.2923 | Yes |
| 96 | Sgms2 | 14038 | -0.183 | -0.2931 | Yes |
| 97 | Tnfsf10 | 14053 | -0.183 | -0.2893 | Yes |
| 98 | Csf1 | 14086 | -0.185 | -0.2863 | Yes |
| 99 | Ifitm1 | 14149 | -0.188 | -0.2847 | Yes |
| 100 | Nmi | 14210 | -0.191 | -0.2830 | Yes |
| 101 | Rtp4 | 14261 | -0.193 | -0.2807 | Yes |
| 102 | Mxd1 | 14266 | -0.194 | -0.2761 | Yes |
| 103 | Plaur | 14288 | -0.195 | -0.2723 | Yes |
| 104 | Tacr1 | 14514 | -0.208 | -0.2786 | Yes |
| 105 | Nfkb1 | 14543 | -0.209 | -0.2748 | Yes |
| 106 | Nod2 | 14579 | -0.210 | -0.2713 | Yes |
| 107 | Kif1b | 14759 | -0.219 | -0.2750 | Yes |
| 108 | Myc | 14831 | -0.223 | -0.2730 | Yes |
| 109 | Slamf1 | 14903 | -0.227 | -0.2710 | Yes |
| 110 | Ripk2 | 14923 | -0.228 | -0.2663 | Yes |
| 111 | Gpr183 | 14934 | -0.228 | -0.2611 | Yes |
| 112 | Cx3cl1 | 15075 | -0.234 | -0.2623 | Yes |
| 113 | Irak2 | 15098 | -0.235 | -0.2576 | Yes |
| 114 | Sele | 15192 | -0.239 | -0.2563 | Yes |
| 115 | Kcna3 | 15201 | -0.240 | -0.2507 | Yes |
| 116 | Gabbr1 | 15305 | -0.244 | -0.2499 | Yes |
| 117 | Tnfrsf9 | 15429 | -0.250 | -0.2499 | Yes |
| 118 | Rhog | 15534 | -0.256 | -0.2488 | Yes |
| 119 | Cxcl9 | 15624 | -0.260 | -0.2468 | Yes |
| 120 | Tacr3 | 15689 | -0.264 | -0.2435 | Yes |
| 121 | Slc31a1 | 15718 | -0.265 | -0.2383 | Yes |
| 122 | Emp3 | 15791 | -0.270 | -0.2352 | Yes |
| 123 | Slc28a2 | 15921 | -0.280 | -0.2348 | Yes |
| 124 | Ebi3 | 16016 | -0.287 | -0.2324 | Yes |
| 125 | Lck | 16097 | -0.291 | -0.2292 | Yes |
| 126 | Ptger4 | 16130 | -0.293 | -0.2235 | Yes |
| 127 | Npffr2 | 16399 | -0.310 | -0.2294 | Yes |
| 128 | Kcnmb2 | 16600 | -0.324 | -0.2315 | Yes |
| 129 | Tnfsf15 | 16660 | -0.327 | -0.2264 | Yes |
| 130 | Icam1 | 16779 | -0.335 | -0.2240 | Yes |
| 131 | Tlr2 | 16800 | -0.337 | -0.2166 | Yes |
| 132 | Cd69 | 16936 | -0.347 | -0.2148 | Yes |
| 133 | Scarf1 | 16971 | -0.350 | -0.2078 | Yes |
| 134 | Clec5a | 17056 | -0.355 | -0.2032 | Yes |
| 135 | Il18 | 17107 | -0.359 | -0.1968 | Yes |
| 136 | F3 | 17195 | -0.365 | -0.1921 | Yes |
| 137 | Gpr132 | 17245 | -0.370 | -0.1854 | Yes |
| 138 | Ptger2 | 17666 | -0.405 | -0.1966 | Yes |
| 139 | Calcrl | 17804 | -0.418 | -0.1932 | Yes |
| 140 | Selenos | 17827 | -0.419 | -0.1838 | Yes |
| 141 | Sri | 17838 | -0.420 | -0.1738 | Yes |
| 142 | Ccrl2 | 18120 | -0.446 | -0.1770 | Yes |
| 143 | Il18r1 | 18144 | -0.449 | -0.1669 | Yes |
| 144 | Ly6e | 18147 | -0.449 | -0.1558 | Yes |
| 145 | Adora2b | 18230 | -0.459 | -0.1485 | Yes |
| 146 | Ifngr2 | 18245 | -0.460 | -0.1377 | Yes |
| 147 | Cybb | 18616 | -0.508 | -0.1439 | Yes |
| 148 | Cd14 | 18844 | -0.547 | -0.1418 | Yes |
| 149 | Hpn | 19052 | -0.585 | -0.1377 | Yes |
| 150 | Ptgir | 19078 | -0.591 | -0.1242 | Yes |
| 151 | Il15ra | 19097 | -0.594 | -0.1103 | Yes |
| 152 | P2rx4 | 19225 | -0.626 | -0.1011 | Yes |
| 153 | Nfkbia | 19262 | -0.638 | -0.0870 | Yes |
| 154 | Cd48 | 19319 | -0.662 | -0.0733 | Yes |
| 155 | Ccl5 | 19331 | -0.664 | -0.0573 | Yes |
| 156 | Pvr | 19504 | -0.737 | -0.0476 | Yes |
| 157 | Mefv | 19568 | -0.780 | -0.0313 | Yes |
| 158 | Gch1 | 19570 | -0.782 | -0.0119 | Yes |
| 159 | Tlr1 | 19665 | -0.882 | 0.0054 | Yes |