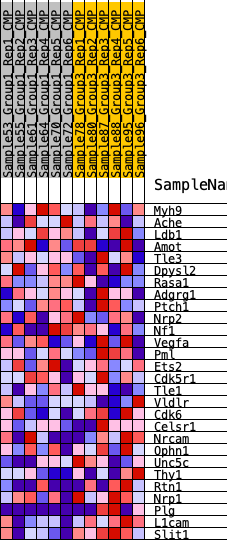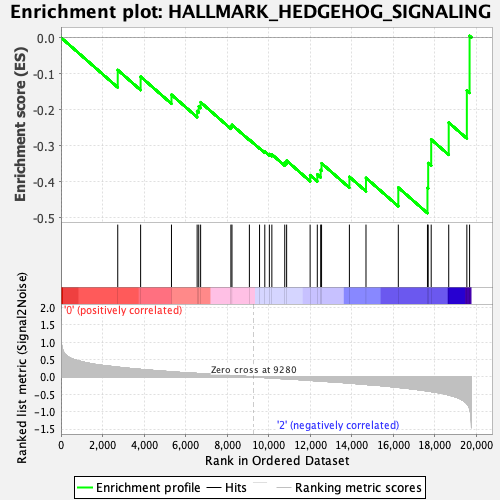
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | -0.486989 |
| Normalized Enrichment Score (NES) | -1.4517785 |
| Nominal p-value | 0.07597536 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.521 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Myh9 | 2733 | 0.279 | -0.0900 | No |
| 2 | Ache | 3835 | 0.215 | -0.1084 | No |
| 3 | Ldb1 | 5321 | 0.145 | -0.1584 | No |
| 4 | Amot | 6557 | 0.099 | -0.2038 | No |
| 5 | Tle3 | 6635 | 0.096 | -0.1910 | No |
| 6 | Dpysl2 | 6725 | 0.093 | -0.1794 | No |
| 7 | Rasa1 | 8183 | 0.040 | -0.2462 | No |
| 8 | Adgrg1 | 8235 | 0.038 | -0.2421 | No |
| 9 | Ptch1 | 9074 | 0.008 | -0.2832 | No |
| 10 | Nrp2 | 9563 | -0.007 | -0.3068 | No |
| 11 | Nf1 | 9816 | -0.018 | -0.3165 | No |
| 12 | Vegfa | 10036 | -0.025 | -0.3231 | No |
| 13 | Pml | 10156 | -0.030 | -0.3240 | No |
| 14 | Ets2 | 10777 | -0.052 | -0.3464 | No |
| 15 | Cdk5r1 | 10868 | -0.055 | -0.3414 | No |
| 16 | Tle1 | 11999 | -0.098 | -0.3817 | No |
| 17 | Vldlr | 12345 | -0.112 | -0.3797 | No |
| 18 | Cdk6 | 12508 | -0.118 | -0.3674 | No |
| 19 | Celsr1 | 12550 | -0.120 | -0.3487 | No |
| 20 | Nrcam | 13891 | -0.175 | -0.3862 | No |
| 21 | Ophn1 | 14690 | -0.216 | -0.3892 | No |
| 22 | Unc5c | 16247 | -0.300 | -0.4159 | No |
| 23 | Thy1 | 17651 | -0.404 | -0.4169 | Yes |
| 24 | Rtn1 | 17686 | -0.407 | -0.3481 | Yes |
| 25 | Nrp1 | 17831 | -0.420 | -0.2826 | Yes |
| 26 | Plg | 18675 | -0.516 | -0.2357 | Yes |
| 27 | L1cam | 19548 | -0.767 | -0.1469 | Yes |
| 28 | Slit1 | 19680 | -0.912 | 0.0046 | Yes |