Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.CMP_Pheno.cls#Group1_versus_Group3.CMP_Pheno.cls#Group1_versus_Group3_repos |
| Phenotype | CMP_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
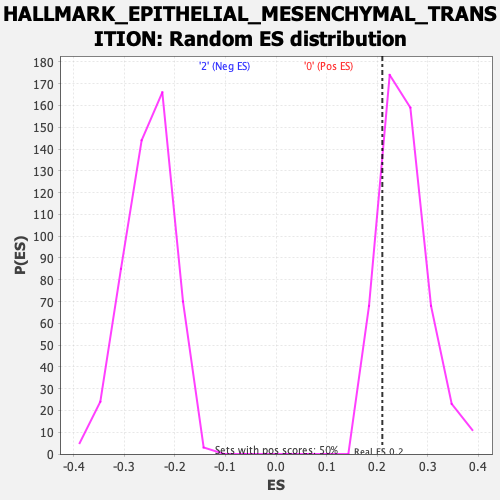
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.21026494 |
| Normalized Enrichment Score (NES) | 0.8314358 |
| Nominal p-value | 0.82306165 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mmp2 | 25 | 0.934 | 0.0247 | Yes |
| 2 | Comp | 29 | 0.919 | 0.0500 | Yes |
| 3 | Tgm2 | 270 | 0.610 | 0.0548 | Yes |
| 4 | Nid2 | 387 | 0.562 | 0.0645 | Yes |
| 5 | Fbn1 | 466 | 0.536 | 0.0754 | Yes |
| 6 | Lamc1 | 512 | 0.525 | 0.0877 | Yes |
| 7 | Dab2 | 568 | 0.509 | 0.0990 | Yes |
| 8 | Thbs1 | 697 | 0.486 | 0.1060 | Yes |
| 9 | Fgf2 | 705 | 0.485 | 0.1191 | Yes |
| 10 | Fstl1 | 723 | 0.481 | 0.1316 | Yes |
| 11 | Sgcb | 1050 | 0.429 | 0.1269 | Yes |
| 12 | Anpep | 1069 | 0.426 | 0.1379 | Yes |
| 13 | Tfpi2 | 1256 | 0.401 | 0.1396 | Yes |
| 14 | Slit2 | 1275 | 0.400 | 0.1497 | Yes |
| 15 | Copa | 1317 | 0.396 | 0.1586 | Yes |
| 16 | Col5a3 | 1407 | 0.382 | 0.1647 | Yes |
| 17 | Postn | 1506 | 0.372 | 0.1701 | Yes |
| 18 | Dpysl3 | 1814 | 0.344 | 0.1640 | Yes |
| 19 | Il6 | 1933 | 0.335 | 0.1673 | Yes |
| 20 | Thbs2 | 1952 | 0.334 | 0.1757 | Yes |
| 21 | Rhob | 1957 | 0.333 | 0.1847 | Yes |
| 22 | Itgb1 | 2053 | 0.326 | 0.1889 | Yes |
| 23 | Dst | 2155 | 0.318 | 0.1926 | Yes |
| 24 | Sdc1 | 2294 | 0.309 | 0.1942 | Yes |
| 25 | Fn1 | 2385 | 0.303 | 0.1980 | Yes |
| 26 | Fas | 2636 | 0.288 | 0.1933 | Yes |
| 27 | Calu | 2668 | 0.285 | 0.1996 | Yes |
| 28 | Jun | 2674 | 0.284 | 0.2073 | Yes |
| 29 | Mmp14 | 2995 | 0.263 | 0.1983 | Yes |
| 30 | Mcm7 | 3065 | 0.259 | 0.2020 | Yes |
| 31 | Timp3 | 3164 | 0.254 | 0.2040 | Yes |
| 32 | Tnc | 3180 | 0.252 | 0.2103 | Yes |
| 33 | P3h1 | 3462 | 0.235 | 0.2025 | No |
| 34 | Fbln1 | 3614 | 0.227 | 0.2011 | No |
| 35 | Basp1 | 3644 | 0.225 | 0.2059 | No |
| 36 | Cap2 | 3846 | 0.215 | 0.2016 | No |
| 37 | Fzd8 | 3909 | 0.212 | 0.2043 | No |
| 38 | Notch2 | 3918 | 0.211 | 0.2098 | No |
| 39 | Tpm2 | 4310 | 0.193 | 0.1952 | No |
| 40 | Gja1 | 4785 | 0.171 | 0.1759 | No |
| 41 | Sparc | 4824 | 0.169 | 0.1786 | No |
| 42 | Plod2 | 4940 | 0.163 | 0.1773 | No |
| 43 | Tnfrsf12a | 5209 | 0.151 | 0.1678 | No |
| 44 | Flna | 5404 | 0.141 | 0.1619 | No |
| 45 | Edil3 | 5504 | 0.137 | 0.1606 | No |
| 46 | Spp1 | 5670 | 0.131 | 0.1559 | No |
| 47 | Nt5e | 5800 | 0.126 | 0.1528 | No |
| 48 | Col1a2 | 5858 | 0.124 | 0.1533 | No |
| 49 | Tgfbi | 5936 | 0.121 | 0.1528 | No |
| 50 | Sfrp4 | 6299 | 0.110 | 0.1374 | No |
| 51 | Il15 | 6510 | 0.101 | 0.1295 | No |
| 52 | Lrp1 | 6795 | 0.090 | 0.1175 | No |
| 53 | Cdh2 | 7012 | 0.083 | 0.1088 | No |
| 54 | Vcam1 | 7842 | 0.052 | 0.0681 | No |
| 55 | Fap | 8099 | 0.043 | 0.0562 | No |
| 56 | Itgb3 | 8227 | 0.038 | 0.0508 | No |
| 57 | Adam12 | 8283 | 0.037 | 0.0491 | No |
| 58 | Itga2 | 8888 | 0.014 | 0.0187 | No |
| 59 | Col4a2 | 9031 | 0.009 | 0.0117 | No |
| 60 | Efemp2 | 9032 | 0.009 | 0.0120 | No |
| 61 | Tgfbr3 | 9180 | 0.004 | 0.0046 | No |
| 62 | Col1a1 | 9280 | 0.000 | -0.0004 | No |
| 63 | Matn2 | 9619 | -0.009 | -0.0174 | No |
| 64 | Pcolce | 9760 | -0.016 | -0.0241 | No |
| 65 | Cd44 | 9782 | -0.016 | -0.0247 | No |
| 66 | Id2 | 9795 | -0.017 | -0.0249 | No |
| 67 | Sgcd | 9801 | -0.017 | -0.0246 | No |
| 68 | Cadm1 | 9893 | -0.020 | -0.0287 | No |
| 69 | Sat1 | 9948 | -0.022 | -0.0308 | No |
| 70 | Vegfa | 10036 | -0.025 | -0.0346 | No |
| 71 | Pdgfrb | 10086 | -0.027 | -0.0363 | No |
| 72 | Wipf1 | 10100 | -0.028 | -0.0362 | No |
| 73 | Pdlim4 | 10432 | -0.040 | -0.0519 | No |
| 74 | Dcn | 10434 | -0.040 | -0.0509 | No |
| 75 | Bgn | 10645 | -0.047 | -0.0603 | No |
| 76 | Fbn2 | 10704 | -0.049 | -0.0618 | No |
| 77 | Gem | 10926 | -0.057 | -0.0715 | No |
| 78 | Qsox1 | 11080 | -0.060 | -0.0776 | No |
| 79 | Tagln | 11373 | -0.072 | -0.0905 | No |
| 80 | Ntm | 11774 | -0.088 | -0.1084 | No |
| 81 | Gpc1 | 11964 | -0.096 | -0.1154 | No |
| 82 | Itgav | 11973 | -0.097 | -0.1131 | No |
| 83 | Gpx7 | 12012 | -0.099 | -0.1123 | No |
| 84 | Lama2 | 12045 | -0.100 | -0.1112 | No |
| 85 | Capg | 12100 | -0.102 | -0.1111 | No |
| 86 | Plod1 | 12254 | -0.108 | -0.1159 | No |
| 87 | Col5a1 | 12554 | -0.120 | -0.1277 | No |
| 88 | Col4a1 | 12586 | -0.121 | -0.1260 | No |
| 89 | Itgb5 | 12598 | -0.122 | -0.1231 | No |
| 90 | Lama1 | 12644 | -0.124 | -0.1220 | No |
| 91 | Slc6a8 | 12976 | -0.137 | -0.1350 | No |
| 92 | Loxl1 | 13037 | -0.139 | -0.1342 | No |
| 93 | Pfn2 | 13083 | -0.139 | -0.1326 | No |
| 94 | Tpm1 | 13299 | -0.149 | -0.1394 | No |
| 95 | Ppib | 13402 | -0.153 | -0.1404 | No |
| 96 | Inhba | 13733 | -0.168 | -0.1525 | No |
| 97 | Itga5 | 13811 | -0.172 | -0.1517 | No |
| 98 | Ecm1 | 13908 | -0.176 | -0.1517 | No |
| 99 | Pmepa1 | 13933 | -0.177 | -0.1480 | No |
| 100 | Sntb1 | 13947 | -0.178 | -0.1437 | No |
| 101 | Cald1 | 13995 | -0.181 | -0.1410 | No |
| 102 | Plaur | 14288 | -0.195 | -0.1505 | No |
| 103 | Colgalt1 | 14630 | -0.213 | -0.1620 | No |
| 104 | Col12a1 | 15246 | -0.242 | -0.1866 | No |
| 105 | Fstl3 | 15396 | -0.249 | -0.1873 | No |
| 106 | Mest | 15763 | -0.268 | -0.1984 | No |
| 107 | Emp3 | 15791 | -0.270 | -0.1923 | No |
| 108 | Plod3 | 15838 | -0.274 | -0.1870 | No |
| 109 | Glipr1 | 16117 | -0.292 | -0.1931 | No |
| 110 | Slit3 | 16315 | -0.305 | -0.1946 | No |
| 111 | Mylk | 16336 | -0.306 | -0.1871 | No |
| 112 | Col16a1 | 16505 | -0.317 | -0.1869 | No |
| 113 | Vegfc | 16640 | -0.327 | -0.1847 | No |
| 114 | Tgfb1 | 16909 | -0.345 | -0.1887 | No |
| 115 | Lama3 | 17036 | -0.354 | -0.1853 | No |
| 116 | Igfbp4 | 17206 | -0.366 | -0.1837 | No |
| 117 | Vcan | 17322 | -0.376 | -0.1791 | No |
| 118 | Fuca1 | 17404 | -0.383 | -0.1726 | No |
| 119 | Bmp1 | 17557 | -0.395 | -0.1694 | No |
| 120 | Thy1 | 17651 | -0.404 | -0.1629 | No |
| 121 | Tpm4 | 17655 | -0.404 | -0.1518 | No |
| 122 | Cdh11 | 17663 | -0.405 | -0.1409 | No |
| 123 | Ecm2 | 17911 | -0.427 | -0.1416 | No |
| 124 | Magee1 | 17934 | -0.429 | -0.1308 | No |
| 125 | Col5a2 | 17993 | -0.434 | -0.1217 | No |
| 126 | Eno2 | 18034 | -0.440 | -0.1115 | No |
| 127 | Spock1 | 18056 | -0.441 | -0.1003 | No |
| 128 | Vim | 18155 | -0.450 | -0.0928 | No |
| 129 | Sfrp1 | 18182 | -0.454 | -0.0815 | No |
| 130 | Serpine2 | 18354 | -0.472 | -0.0771 | No |
| 131 | Abi3bp | 18564 | -0.502 | -0.0738 | No |
| 132 | Lgals1 | 18742 | -0.530 | -0.0681 | No |
| 133 | Gadd45b | 18787 | -0.537 | -0.0554 | No |
| 134 | Gadd45a | 19302 | -0.655 | -0.0634 | No |
| 135 | Tnfaip3 | 19425 | -0.702 | -0.0501 | No |
| 136 | Pvr | 19504 | -0.737 | -0.0336 | No |
| 137 | Col7a1 | 19571 | -0.782 | -0.0152 | No |
| 138 | Serpinh1 | 19681 | -0.913 | 0.0046 | No |