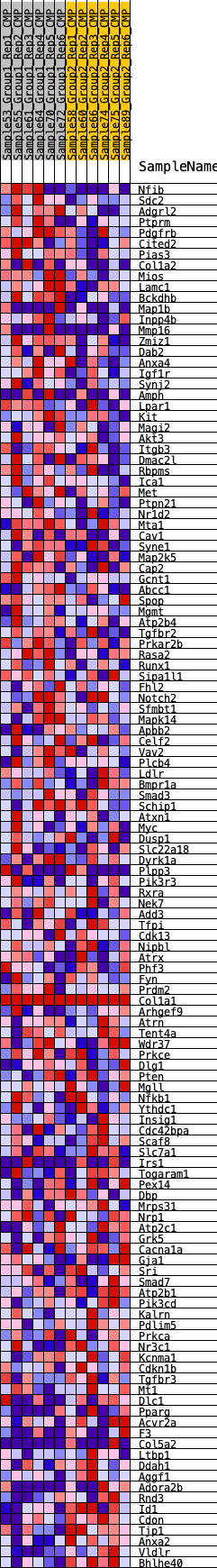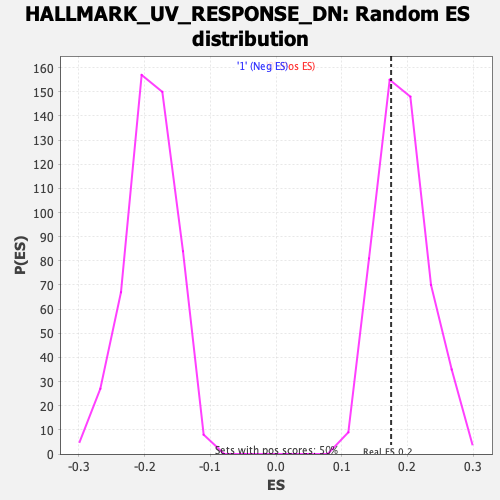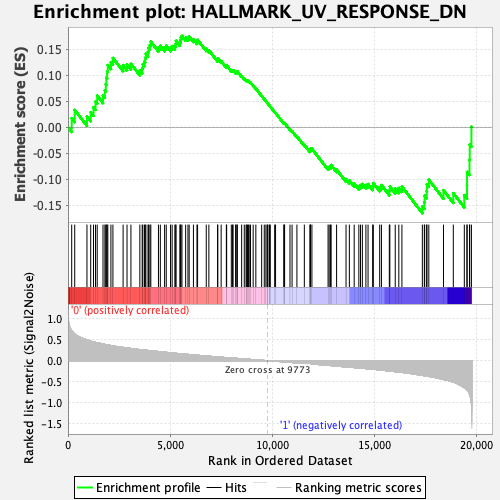
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UV_RESPONSE_DN |
| Enrichment Score (ES) | 0.17521062 |
| Normalized Enrichment Score (NES) | 0.91428506 |
| Nominal p-value | 0.6075697 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nfib | 181 | 0.719 | 0.0173 | Yes |
| 2 | Sdc2 | 329 | 0.630 | 0.0331 | Yes |
| 3 | Adgrl2 | 928 | 0.494 | 0.0208 | Yes |
| 4 | Ptprm | 1112 | 0.464 | 0.0286 | Yes |
| 5 | Pdgfrb | 1248 | 0.445 | 0.0382 | Yes |
| 6 | Cited2 | 1348 | 0.434 | 0.0491 | Yes |
| 7 | Pias3 | 1430 | 0.425 | 0.0607 | Yes |
| 8 | Col1a2 | 1714 | 0.396 | 0.0609 | Yes |
| 9 | Mios | 1806 | 0.386 | 0.0705 | Yes |
| 10 | Lamc1 | 1851 | 0.383 | 0.0824 | Yes |
| 11 | Bckdhb | 1877 | 0.380 | 0.0951 | Yes |
| 12 | Map1b | 1917 | 0.376 | 0.1070 | Yes |
| 13 | Inpp4b | 1950 | 0.374 | 0.1192 | Yes |
| 14 | Mmp16 | 2101 | 0.359 | 0.1248 | Yes |
| 15 | Zmiz1 | 2199 | 0.351 | 0.1328 | Yes |
| 16 | Dab2 | 2698 | 0.315 | 0.1191 | Yes |
| 17 | Anxa4 | 2890 | 0.303 | 0.1205 | Yes |
| 18 | Igf1r | 3081 | 0.290 | 0.1215 | Yes |
| 19 | Synj2 | 3518 | 0.263 | 0.1091 | Yes |
| 20 | Amph | 3632 | 0.257 | 0.1128 | Yes |
| 21 | Lpar1 | 3663 | 0.256 | 0.1207 | Yes |
| 22 | Kit | 3744 | 0.252 | 0.1259 | Yes |
| 23 | Magi2 | 3782 | 0.250 | 0.1332 | Yes |
| 24 | Akt3 | 3813 | 0.248 | 0.1409 | Yes |
| 25 | Itgb3 | 3917 | 0.243 | 0.1446 | Yes |
| 26 | Dmac2l | 3946 | 0.241 | 0.1521 | Yes |
| 27 | Rbpms | 4010 | 0.239 | 0.1577 | Yes |
| 28 | Ica1 | 4055 | 0.236 | 0.1641 | Yes |
| 29 | Met | 4429 | 0.217 | 0.1532 | Yes |
| 30 | Ptpn21 | 4521 | 0.212 | 0.1564 | Yes |
| 31 | Nr1d2 | 4730 | 0.202 | 0.1532 | Yes |
| 32 | Mta1 | 4810 | 0.198 | 0.1565 | Yes |
| 33 | Cav1 | 5020 | 0.188 | 0.1528 | Yes |
| 34 | Syne1 | 5099 | 0.184 | 0.1556 | Yes |
| 35 | Map2k5 | 5232 | 0.178 | 0.1554 | Yes |
| 36 | Cap2 | 5276 | 0.176 | 0.1597 | Yes |
| 37 | Gcnt1 | 5286 | 0.175 | 0.1657 | Yes |
| 38 | Abcc1 | 5478 | 0.166 | 0.1621 | Yes |
| 39 | Spop | 5503 | 0.165 | 0.1670 | Yes |
| 40 | Mgmt | 5529 | 0.164 | 0.1718 | Yes |
| 41 | Atp2b4 | 5579 | 0.161 | 0.1752 | Yes |
| 42 | Tgfbr2 | 5760 | 0.154 | 0.1717 | No |
| 43 | Prkar2b | 5874 | 0.149 | 0.1715 | No |
| 44 | Rasa2 | 5932 | 0.146 | 0.1739 | No |
| 45 | Runx1 | 6141 | 0.136 | 0.1684 | No |
| 46 | Sipa1l1 | 6315 | 0.129 | 0.1644 | No |
| 47 | Fhl2 | 6345 | 0.128 | 0.1676 | No |
| 48 | Notch2 | 6769 | 0.112 | 0.1502 | No |
| 49 | Sfmbt1 | 6897 | 0.106 | 0.1476 | No |
| 50 | Mapk14 | 7325 | 0.089 | 0.1292 | No |
| 51 | Apbb2 | 7332 | 0.089 | 0.1322 | No |
| 52 | Celf2 | 7498 | 0.082 | 0.1268 | No |
| 53 | Vav2 | 7753 | 0.073 | 0.1166 | No |
| 54 | Plcb4 | 7768 | 0.073 | 0.1185 | No |
| 55 | Ldlr | 7996 | 0.064 | 0.1093 | No |
| 56 | Bmpr1a | 8052 | 0.062 | 0.1088 | No |
| 57 | Smad3 | 8083 | 0.060 | 0.1095 | No |
| 58 | Schip1 | 8207 | 0.056 | 0.1053 | No |
| 59 | Atxn1 | 8228 | 0.055 | 0.1063 | No |
| 60 | Myc | 8283 | 0.054 | 0.1056 | No |
| 61 | Dusp1 | 8286 | 0.054 | 0.1075 | No |
| 62 | Slc22a18 | 8502 | 0.045 | 0.0982 | No |
| 63 | Dyrk1a | 8631 | 0.041 | 0.0932 | No |
| 64 | Plpp3 | 8718 | 0.038 | 0.0902 | No |
| 65 | Pik3r3 | 8749 | 0.037 | 0.0901 | No |
| 66 | Rxra | 8790 | 0.036 | 0.0893 | No |
| 67 | Nek7 | 8834 | 0.034 | 0.0884 | No |
| 68 | Add3 | 8870 | 0.032 | 0.0878 | No |
| 69 | Tfpi | 8936 | 0.030 | 0.0856 | No |
| 70 | Cdk13 | 9066 | 0.026 | 0.0800 | No |
| 71 | Nipbl | 9197 | 0.021 | 0.0741 | No |
| 72 | Atrx | 9482 | 0.011 | 0.0601 | No |
| 73 | Phf3 | 9610 | 0.006 | 0.0538 | No |
| 74 | Fyn | 9689 | 0.003 | 0.0500 | No |
| 75 | Prdm2 | 9751 | 0.001 | 0.0469 | No |
| 76 | Col1a1 | 9773 | 0.000 | 0.0458 | No |
| 77 | Arhgef9 | 9845 | -0.001 | 0.0422 | No |
| 78 | Atrn | 9868 | -0.002 | 0.0412 | No |
| 79 | Tent4a | 9904 | -0.003 | 0.0395 | No |
| 80 | Wdr37 | 10119 | -0.011 | 0.0290 | No |
| 81 | Prkce | 10162 | -0.013 | 0.0274 | No |
| 82 | Dlg1 | 10566 | -0.027 | 0.0078 | No |
| 83 | Pten | 10586 | -0.027 | 0.0079 | No |
| 84 | Mgll | 10605 | -0.028 | 0.0080 | No |
| 85 | Nfkb1 | 10869 | -0.038 | -0.0040 | No |
| 86 | Ythdc1 | 10971 | -0.042 | -0.0076 | No |
| 87 | Insig1 | 11208 | -0.051 | -0.0177 | No |
| 88 | Cdc42bpa | 11571 | -0.063 | -0.0338 | No |
| 89 | Scaf8 | 11844 | -0.073 | -0.0449 | No |
| 90 | Slc7a1 | 11866 | -0.074 | -0.0432 | No |
| 91 | Irs1 | 11873 | -0.075 | -0.0408 | No |
| 92 | Togaram1 | 11942 | -0.078 | -0.0414 | No |
| 93 | Pex14 | 12733 | -0.109 | -0.0776 | No |
| 94 | Dbp | 12787 | -0.111 | -0.0762 | No |
| 95 | Mrps31 | 12855 | -0.114 | -0.0754 | No |
| 96 | Nrp1 | 12882 | -0.115 | -0.0725 | No |
| 97 | Atp2c1 | 13151 | -0.127 | -0.0814 | No |
| 98 | Grk5 | 13613 | -0.145 | -0.0995 | No |
| 99 | Cacna1a | 13776 | -0.152 | -0.1021 | No |
| 100 | Gja1 | 14012 | -0.163 | -0.1081 | No |
| 101 | Sri | 14240 | -0.172 | -0.1133 | No |
| 102 | Smad7 | 14322 | -0.176 | -0.1110 | No |
| 103 | Atp2b1 | 14412 | -0.179 | -0.1089 | No |
| 104 | Pik3cd | 14585 | -0.187 | -0.1108 | No |
| 105 | Kalrn | 14691 | -0.191 | -0.1091 | No |
| 106 | Pdlim5 | 14916 | -0.201 | -0.1130 | No |
| 107 | Prkca | 14948 | -0.203 | -0.1071 | No |
| 108 | Nr3c1 | 15261 | -0.219 | -0.1149 | No |
| 109 | Kcnma1 | 15346 | -0.223 | -0.1110 | No |
| 110 | Cdkn1b | 15732 | -0.244 | -0.1216 | No |
| 111 | Tgfbr3 | 15758 | -0.246 | -0.1138 | No |
| 112 | Mt1 | 16022 | -0.260 | -0.1176 | No |
| 113 | Dlc1 | 16197 | -0.269 | -0.1165 | No |
| 114 | Pparg | 16349 | -0.277 | -0.1140 | No |
| 115 | Acvr2a | 17349 | -0.349 | -0.1520 | No |
| 116 | F3 | 17446 | -0.357 | -0.1437 | No |
| 117 | Col5a2 | 17463 | -0.358 | -0.1313 | No |
| 118 | Ltbp1 | 17562 | -0.365 | -0.1228 | No |
| 119 | Ddah1 | 17572 | -0.366 | -0.1098 | No |
| 120 | Aggf1 | 17666 | -0.374 | -0.1007 | No |
| 121 | Adora2b | 18384 | -0.445 | -0.1208 | No |
| 122 | Rnd3 | 18865 | -0.508 | -0.1265 | No |
| 123 | Id1 | 19404 | -0.641 | -0.1303 | No |
| 124 | Cdon | 19535 | -0.693 | -0.1113 | No |
| 125 | Tjp1 | 19539 | -0.695 | -0.0858 | No |
| 126 | Anxa2 | 19657 | -0.790 | -0.0626 | No |
| 127 | Vldlr | 19669 | -0.812 | -0.0333 | No |
| 128 | Bhlhe40 | 19752 | -1.041 | 0.0010 | No |