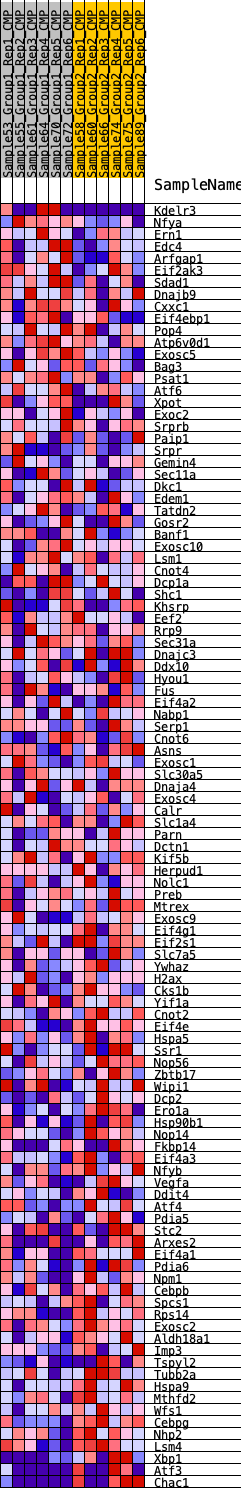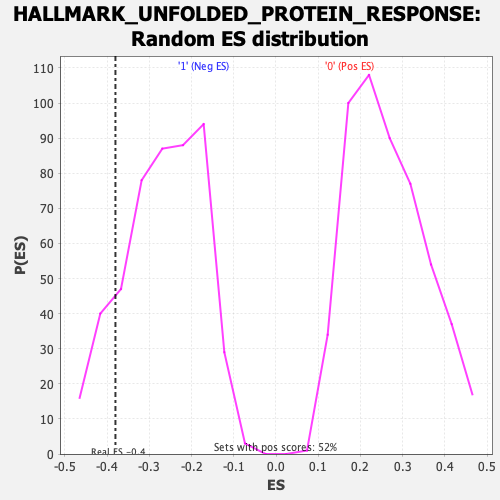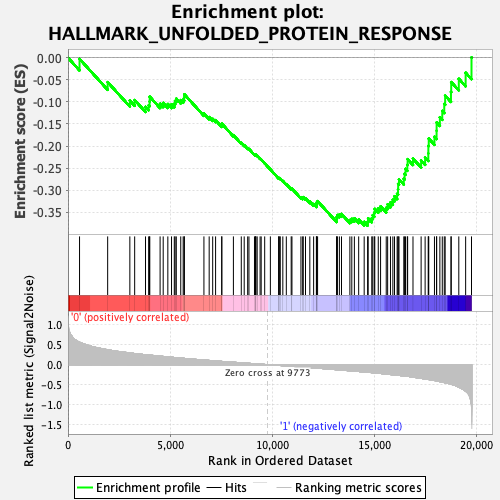
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.38029975 |
| Normalized Enrichment Score (NES) | -1.4282898 |
| Nominal p-value | 0.1307054 |
| FDR q-value | 0.24954695 |
| FWER p-Value | 0.546 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Kdelr3 | 565 | 0.562 | -0.0027 | No |
| 2 | Nfya | 1942 | 0.375 | -0.0553 | No |
| 3 | Ern1 | 3028 | 0.294 | -0.0969 | No |
| 4 | Edc4 | 3264 | 0.279 | -0.0959 | No |
| 5 | Arfgap1 | 3795 | 0.250 | -0.1113 | No |
| 6 | Eif2ak3 | 3949 | 0.241 | -0.1079 | No |
| 7 | Sdad1 | 3995 | 0.240 | -0.0991 | No |
| 8 | Dnajb9 | 4001 | 0.239 | -0.0883 | No |
| 9 | Cxxc1 | 4509 | 0.213 | -0.1042 | No |
| 10 | Eif4ebp1 | 4661 | 0.206 | -0.1023 | No |
| 11 | Pop4 | 4888 | 0.194 | -0.1048 | No |
| 12 | Atp6v0d1 | 5069 | 0.186 | -0.1054 | No |
| 13 | Exosc5 | 5204 | 0.178 | -0.1039 | No |
| 14 | Bag3 | 5244 | 0.177 | -0.0977 | No |
| 15 | Psat1 | 5302 | 0.174 | -0.0926 | No |
| 16 | Atf6 | 5520 | 0.164 | -0.0960 | No |
| 17 | Xpot | 5639 | 0.159 | -0.0946 | No |
| 18 | Exoc2 | 5690 | 0.156 | -0.0899 | No |
| 19 | Srprb | 5695 | 0.156 | -0.0829 | No |
| 20 | Paip1 | 6655 | 0.115 | -0.1263 | No |
| 21 | Srpr | 6912 | 0.105 | -0.1345 | No |
| 22 | Gemin4 | 7084 | 0.099 | -0.1386 | No |
| 23 | Sec11a | 7226 | 0.093 | -0.1414 | No |
| 24 | Dkc1 | 7528 | 0.081 | -0.1530 | No |
| 25 | Edem1 | 7530 | 0.081 | -0.1493 | No |
| 26 | Tatdn2 | 8098 | 0.060 | -0.1754 | No |
| 27 | Gosr2 | 8484 | 0.046 | -0.1928 | No |
| 28 | Banf1 | 8627 | 0.041 | -0.1981 | No |
| 29 | Exosc10 | 8797 | 0.035 | -0.2051 | No |
| 30 | Lsm1 | 8859 | 0.033 | -0.2067 | No |
| 31 | Cnot4 | 9130 | 0.023 | -0.2193 | No |
| 32 | Dcp1a | 9176 | 0.022 | -0.2206 | No |
| 33 | Shc1 | 9177 | 0.022 | -0.2196 | No |
| 34 | Khsrp | 9204 | 0.020 | -0.2200 | No |
| 35 | Eef2 | 9262 | 0.019 | -0.2220 | No |
| 36 | Rrp9 | 9405 | 0.014 | -0.2286 | No |
| 37 | Sec31a | 9458 | 0.011 | -0.2307 | No |
| 38 | Dnajc3 | 9630 | 0.005 | -0.2391 | No |
| 39 | Ddx10 | 9908 | -0.003 | -0.2531 | No |
| 40 | Hyou1 | 10315 | -0.018 | -0.2729 | No |
| 41 | Fus | 10318 | -0.018 | -0.2722 | No |
| 42 | Eif4a2 | 10364 | -0.020 | -0.2735 | No |
| 43 | Nabp1 | 10391 | -0.020 | -0.2739 | No |
| 44 | Serp1 | 10518 | -0.025 | -0.2792 | No |
| 45 | Cnot6 | 10689 | -0.031 | -0.2864 | No |
| 46 | Asns | 10927 | -0.041 | -0.2965 | No |
| 47 | Exosc1 | 10965 | -0.042 | -0.2965 | No |
| 48 | Slc30a5 | 11405 | -0.058 | -0.3161 | No |
| 49 | Dnaja4 | 11491 | -0.062 | -0.3176 | No |
| 50 | Exosc4 | 11516 | -0.063 | -0.3159 | No |
| 51 | Calr | 11628 | -0.065 | -0.3185 | No |
| 52 | Slc1a4 | 11840 | -0.073 | -0.3258 | No |
| 53 | Parn | 12026 | -0.082 | -0.3315 | No |
| 54 | Dctn1 | 12161 | -0.087 | -0.3343 | No |
| 55 | Kif5b | 12166 | -0.087 | -0.3304 | No |
| 56 | Herpud1 | 12185 | -0.087 | -0.3273 | No |
| 57 | Nolc1 | 12220 | -0.089 | -0.3249 | No |
| 58 | Preb | 13152 | -0.127 | -0.3664 | No |
| 59 | Mtrex | 13159 | -0.127 | -0.3608 | No |
| 60 | Exosc9 | 13190 | -0.128 | -0.3564 | No |
| 61 | Eif4g1 | 13287 | -0.133 | -0.3551 | No |
| 62 | Eif2s1 | 13391 | -0.138 | -0.3539 | No |
| 63 | Slc7a5 | 13801 | -0.154 | -0.3676 | No |
| 64 | Ywhaz | 13895 | -0.157 | -0.3651 | No |
| 65 | H2ax | 14021 | -0.163 | -0.3639 | No |
| 66 | Cks1b | 14232 | -0.171 | -0.3666 | No |
| 67 | Yif1a | 14501 | -0.183 | -0.3718 | Yes |
| 68 | Cnot2 | 14670 | -0.190 | -0.3715 | Yes |
| 69 | Eif4e | 14696 | -0.191 | -0.3639 | Yes |
| 70 | Hspa5 | 14873 | -0.199 | -0.3637 | Yes |
| 71 | Ssr1 | 14921 | -0.202 | -0.3567 | Yes |
| 72 | Nop56 | 14995 | -0.206 | -0.3509 | Yes |
| 73 | Zbtb17 | 15020 | -0.207 | -0.3425 | Yes |
| 74 | Wipi1 | 15191 | -0.215 | -0.3412 | Yes |
| 75 | Dcp2 | 15304 | -0.221 | -0.3367 | Yes |
| 76 | Ero1a | 15581 | -0.236 | -0.3398 | Yes |
| 77 | Hsp90b1 | 15650 | -0.240 | -0.3321 | Yes |
| 78 | Nop14 | 15789 | -0.248 | -0.3277 | Yes |
| 79 | Fkbp14 | 15905 | -0.254 | -0.3218 | Yes |
| 80 | Eif4a3 | 15985 | -0.258 | -0.3138 | Yes |
| 81 | Nfyb | 16115 | -0.265 | -0.3081 | Yes |
| 82 | Vegfa | 16159 | -0.267 | -0.2979 | Yes |
| 83 | Ddit4 | 16165 | -0.267 | -0.2858 | Yes |
| 84 | Atf4 | 16206 | -0.269 | -0.2754 | Yes |
| 85 | Pdia5 | 16441 | -0.283 | -0.2742 | Yes |
| 86 | Stc2 | 16480 | -0.285 | -0.2629 | Yes |
| 87 | Arxes2 | 16526 | -0.288 | -0.2519 | Yes |
| 88 | Eif4a1 | 16613 | -0.293 | -0.2427 | Yes |
| 89 | Pdia6 | 16629 | -0.294 | -0.2298 | Yes |
| 90 | Npm1 | 16891 | -0.313 | -0.2286 | Yes |
| 91 | Cebpb | 17289 | -0.344 | -0.2328 | Yes |
| 92 | Spcs1 | 17485 | -0.360 | -0.2260 | Yes |
| 93 | Rps14 | 17641 | -0.371 | -0.2167 | Yes |
| 94 | Exosc2 | 17644 | -0.372 | -0.1996 | Yes |
| 95 | Aldh18a1 | 17663 | -0.373 | -0.1832 | Yes |
| 96 | Imp3 | 17942 | -0.397 | -0.1789 | Yes |
| 97 | Tspyl2 | 18046 | -0.407 | -0.1653 | Yes |
| 98 | Tubb2a | 18049 | -0.407 | -0.1466 | Yes |
| 99 | Hspa9 | 18207 | -0.426 | -0.1348 | Yes |
| 100 | Mthfd2 | 18325 | -0.438 | -0.1205 | Yes |
| 101 | Wfs1 | 18422 | -0.450 | -0.1045 | Yes |
| 102 | Cebpg | 18466 | -0.454 | -0.0857 | Yes |
| 103 | Nhp2 | 18751 | -0.490 | -0.0774 | Yes |
| 104 | Lsm4 | 18763 | -0.492 | -0.0552 | Yes |
| 105 | Xbp1 | 19134 | -0.566 | -0.0478 | Yes |
| 106 | Atf3 | 19469 | -0.665 | -0.0340 | Yes |
| 107 | Chac1 | 19756 | -1.064 | 0.0008 | Yes |