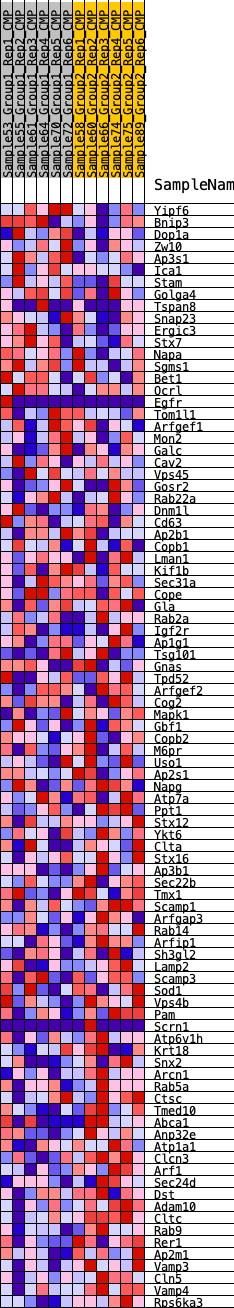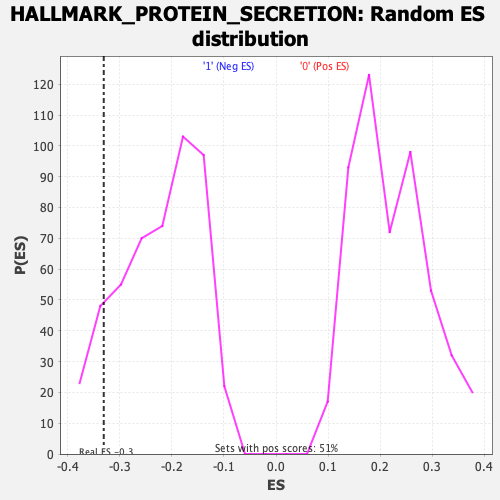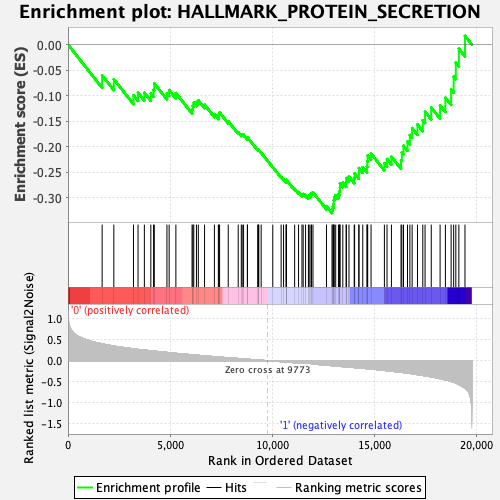
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.33046943 |
| Normalized Enrichment Score (NES) | -1.4873052 |
| Nominal p-value | 0.12398374 |
| FDR q-value | 0.23106709 |
| FWER p-Value | 0.448 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Yipf6 | 1671 | 0.399 | -0.0603 | No |
| 2 | Bnip3 | 2244 | 0.347 | -0.0680 | No |
| 3 | Dop1a | 3206 | 0.282 | -0.0994 | No |
| 4 | Zw10 | 3429 | 0.268 | -0.0942 | No |
| 5 | Ap3s1 | 3739 | 0.252 | -0.0943 | No |
| 6 | Ica1 | 4055 | 0.236 | -0.0958 | No |
| 7 | Stam | 4189 | 0.229 | -0.0884 | No |
| 8 | Golga4 | 4224 | 0.228 | -0.0761 | No |
| 9 | Tspan8 | 4844 | 0.196 | -0.0954 | No |
| 10 | Snap23 | 4952 | 0.190 | -0.0891 | No |
| 11 | Ergic3 | 5282 | 0.175 | -0.0950 | No |
| 12 | Stx7 | 6074 | 0.139 | -0.1266 | No |
| 13 | Napa | 6113 | 0.138 | -0.1201 | No |
| 14 | Sgms1 | 6158 | 0.136 | -0.1139 | No |
| 15 | Bet1 | 6289 | 0.130 | -0.1125 | No |
| 16 | Ocrl | 6381 | 0.127 | -0.1093 | No |
| 17 | Egfr | 6686 | 0.114 | -0.1178 | No |
| 18 | Tom1l1 | 7168 | 0.095 | -0.1363 | No |
| 19 | Arfgef1 | 7361 | 0.088 | -0.1407 | No |
| 20 | Mon2 | 7386 | 0.087 | -0.1366 | No |
| 21 | Galc | 7422 | 0.086 | -0.1331 | No |
| 22 | Cav2 | 7845 | 0.070 | -0.1502 | No |
| 23 | Vps45 | 8333 | 0.052 | -0.1717 | No |
| 24 | Gosr2 | 8484 | 0.046 | -0.1765 | No |
| 25 | Rab22a | 8527 | 0.045 | -0.1759 | No |
| 26 | Dnm1l | 8596 | 0.042 | -0.1767 | No |
| 27 | Cd63 | 8782 | 0.036 | -0.1839 | No |
| 28 | Ap2b1 | 8787 | 0.036 | -0.1819 | No |
| 29 | Copb1 | 9287 | 0.018 | -0.2062 | No |
| 30 | Lman1 | 9330 | 0.017 | -0.2073 | No |
| 31 | Kif1b | 9340 | 0.016 | -0.2067 | No |
| 32 | Sec31a | 9458 | 0.011 | -0.2120 | No |
| 33 | Cope | 10026 | -0.007 | -0.2403 | No |
| 34 | Gla | 10435 | -0.023 | -0.2597 | No |
| 35 | Rab2a | 10554 | -0.026 | -0.2640 | No |
| 36 | Igf2r | 10680 | -0.031 | -0.2685 | No |
| 37 | Ap1g1 | 10690 | -0.031 | -0.2670 | No |
| 38 | Tsg101 | 10691 | -0.031 | -0.2651 | No |
| 39 | Gnas | 11097 | -0.047 | -0.2827 | No |
| 40 | Tpd52 | 11288 | -0.054 | -0.2891 | No |
| 41 | Arfgef2 | 11455 | -0.060 | -0.2938 | No |
| 42 | Cog2 | 11514 | -0.063 | -0.2929 | No |
| 43 | Mapk1 | 11632 | -0.065 | -0.2948 | No |
| 44 | Gbf1 | 11771 | -0.071 | -0.2974 | No |
| 45 | Copb2 | 11808 | -0.072 | -0.2948 | No |
| 46 | M6pr | 11895 | -0.076 | -0.2945 | No |
| 47 | Uso1 | 11923 | -0.077 | -0.2911 | No |
| 48 | Ap2s1 | 11994 | -0.080 | -0.2897 | No |
| 49 | Napg | 12655 | -0.105 | -0.3167 | No |
| 50 | Atp7a | 12926 | -0.117 | -0.3232 | Yes |
| 51 | Ppt1 | 12967 | -0.118 | -0.3180 | Yes |
| 52 | Stx12 | 13005 | -0.120 | -0.3125 | Yes |
| 53 | Ykt6 | 13011 | -0.120 | -0.3054 | Yes |
| 54 | Clta | 13033 | -0.121 | -0.2990 | Yes |
| 55 | Stx16 | 13107 | -0.124 | -0.2950 | Yes |
| 56 | Ap3b1 | 13242 | -0.131 | -0.2937 | Yes |
| 57 | Sec22b | 13289 | -0.133 | -0.2878 | Yes |
| 58 | Tmx1 | 13313 | -0.134 | -0.2807 | Yes |
| 59 | Scamp1 | 13315 | -0.134 | -0.2725 | Yes |
| 60 | Arfgap3 | 13455 | -0.140 | -0.2709 | Yes |
| 61 | Rab14 | 13618 | -0.146 | -0.2702 | Yes |
| 62 | Arfip1 | 13629 | -0.146 | -0.2617 | Yes |
| 63 | Sh3gl2 | 13757 | -0.151 | -0.2588 | Yes |
| 64 | Lamp2 | 14013 | -0.163 | -0.2617 | Yes |
| 65 | Scamp3 | 14033 | -0.163 | -0.2526 | Yes |
| 66 | Sod1 | 14241 | -0.172 | -0.2526 | Yes |
| 67 | Vps4b | 14244 | -0.172 | -0.2421 | Yes |
| 68 | Pam | 14432 | -0.180 | -0.2405 | Yes |
| 69 | Scrn1 | 14637 | -0.189 | -0.2392 | Yes |
| 70 | Atp6v1h | 14654 | -0.189 | -0.2283 | Yes |
| 71 | Krt18 | 14673 | -0.190 | -0.2175 | Yes |
| 72 | Snx2 | 14838 | -0.198 | -0.2137 | Yes |
| 73 | Arcn1 | 15491 | -0.231 | -0.2325 | Yes |
| 74 | Rab5a | 15620 | -0.238 | -0.2243 | Yes |
| 75 | Ctsc | 15835 | -0.250 | -0.2198 | Yes |
| 76 | Tmed10 | 16305 | -0.276 | -0.2267 | Yes |
| 77 | Abca1 | 16346 | -0.277 | -0.2116 | Yes |
| 78 | Anp32e | 16426 | -0.282 | -0.1982 | Yes |
| 79 | Atp1a1 | 16622 | -0.294 | -0.1900 | Yes |
| 80 | Clcn3 | 16739 | -0.302 | -0.1773 | Yes |
| 81 | Arf1 | 16849 | -0.310 | -0.1638 | Yes |
| 82 | Sec24d | 17111 | -0.330 | -0.1567 | Yes |
| 83 | Dst | 17371 | -0.350 | -0.1482 | Yes |
| 84 | Adam10 | 17482 | -0.360 | -0.1316 | Yes |
| 85 | Cltc | 17785 | -0.383 | -0.1233 | Yes |
| 86 | Rab9 | 18218 | -0.426 | -0.1190 | Yes |
| 87 | Rer1 | 18478 | -0.455 | -0.1041 | Yes |
| 88 | Ap2m1 | 18758 | -0.492 | -0.0879 | Yes |
| 89 | Vamp3 | 18882 | -0.510 | -0.0627 | Yes |
| 90 | Cln5 | 18988 | -0.529 | -0.0355 | Yes |
| 91 | Vamp4 | 19137 | -0.566 | -0.0081 | Yes |
| 92 | Rps6ka3 | 19436 | -0.653 | 0.0170 | Yes |