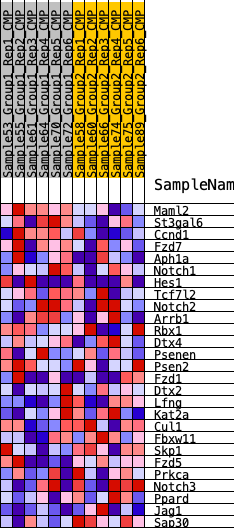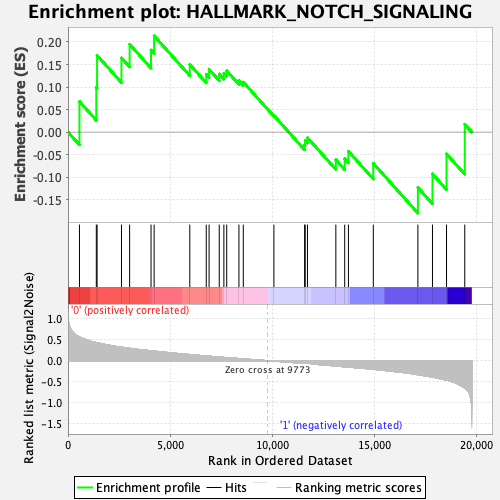
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_NOTCH_SIGNALING |
| Enrichment Score (ES) | 0.21359657 |
| Normalized Enrichment Score (NES) | 0.7183759 |
| Nominal p-value | 0.8842315 |
| FDR q-value | 0.8700962 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Maml2 | 561 | 0.564 | 0.0680 | Yes |
| 2 | St3gal6 | 1386 | 0.429 | 0.0995 | Yes |
| 3 | Ccnd1 | 1424 | 0.426 | 0.1703 | Yes |
| 4 | Fzd7 | 2618 | 0.321 | 0.1648 | Yes |
| 5 | Aph1a | 3017 | 0.294 | 0.1949 | Yes |
| 6 | Notch1 | 4065 | 0.236 | 0.1822 | Yes |
| 7 | Hes1 | 4217 | 0.228 | 0.2136 | Yes |
| 8 | Tcf7l2 | 5963 | 0.145 | 0.1499 | No |
| 9 | Notch2 | 6769 | 0.112 | 0.1283 | No |
| 10 | Arrb1 | 6908 | 0.105 | 0.1393 | No |
| 11 | Rbx1 | 7405 | 0.086 | 0.1289 | No |
| 12 | Dtx4 | 7632 | 0.077 | 0.1306 | No |
| 13 | Psenen | 7771 | 0.072 | 0.1360 | No |
| 14 | Psen2 | 8369 | 0.050 | 0.1143 | No |
| 15 | Fzd1 | 8582 | 0.043 | 0.1109 | No |
| 16 | Dtx2 | 10081 | -0.010 | 0.0367 | No |
| 17 | Lfng | 11586 | -0.064 | -0.0285 | No |
| 18 | Kat2a | 11613 | -0.065 | -0.0188 | No |
| 19 | Cul1 | 11724 | -0.070 | -0.0125 | No |
| 20 | Fbxw11 | 13114 | -0.125 | -0.0615 | No |
| 21 | Skp1 | 13548 | -0.142 | -0.0591 | No |
| 22 | Fzd5 | 13730 | -0.150 | -0.0426 | No |
| 23 | Prkca | 14948 | -0.203 | -0.0694 | No |
| 24 | Notch3 | 17131 | -0.332 | -0.1232 | No |
| 25 | Ppard | 17845 | -0.389 | -0.0928 | No |
| 26 | Jag1 | 18533 | -0.463 | -0.0484 | No |
| 27 | Sap30 | 19426 | -0.650 | 0.0175 | No |