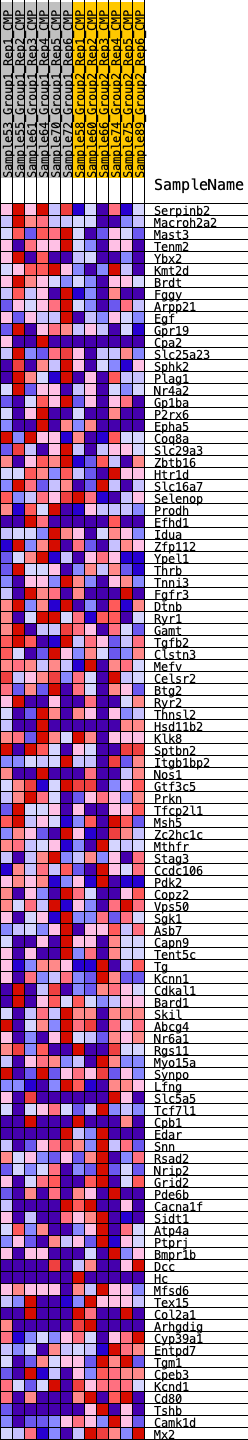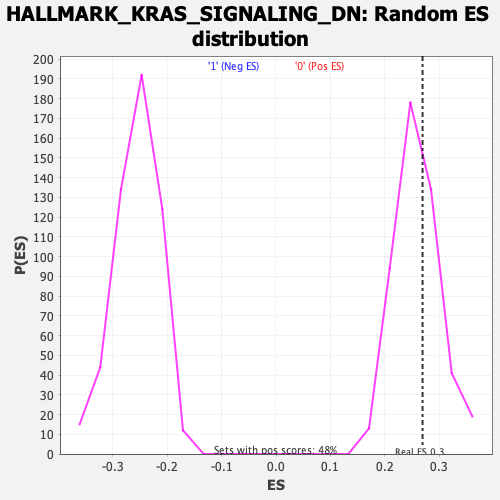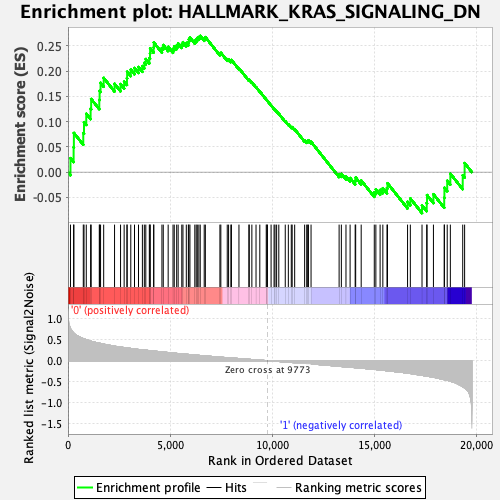
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.26956335 |
| Normalized Enrichment Score (NES) | 1.0422177 |
| Nominal p-value | 0.3716075 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Serpinb2 | 121 | 0.758 | 0.0276 | Yes |
| 2 | Macroh2a2 | 275 | 0.657 | 0.0492 | Yes |
| 3 | Mast3 | 288 | 0.649 | 0.0775 | Yes |
| 4 | Tenm2 | 748 | 0.524 | 0.0775 | Yes |
| 5 | Ybx2 | 784 | 0.519 | 0.0989 | Yes |
| 6 | Kmt2d | 895 | 0.499 | 0.1155 | Yes |
| 7 | Brdt | 1107 | 0.465 | 0.1255 | Yes |
| 8 | Fggy | 1134 | 0.460 | 0.1447 | Yes |
| 9 | Arpp21 | 1532 | 0.415 | 0.1430 | Yes |
| 10 | Egf | 1548 | 0.413 | 0.1607 | Yes |
| 11 | Gpr19 | 1594 | 0.408 | 0.1766 | Yes |
| 12 | Cpa2 | 1747 | 0.393 | 0.1864 | Yes |
| 13 | Slc25a23 | 2279 | 0.344 | 0.1747 | Yes |
| 14 | Sphk2 | 2573 | 0.323 | 0.1742 | Yes |
| 15 | Plag1 | 2749 | 0.311 | 0.1792 | Yes |
| 16 | Nr4a2 | 2886 | 0.303 | 0.1858 | Yes |
| 17 | Gp1ba | 2896 | 0.302 | 0.1988 | Yes |
| 18 | P2rx6 | 3074 | 0.290 | 0.2028 | Yes |
| 19 | Epha5 | 3256 | 0.279 | 0.2060 | Yes |
| 20 | Coq8a | 3452 | 0.266 | 0.2080 | Yes |
| 21 | Slc29a3 | 3642 | 0.257 | 0.2098 | Yes |
| 22 | Zbtb16 | 3737 | 0.252 | 0.2163 | Yes |
| 23 | Htr1d | 3809 | 0.249 | 0.2238 | Yes |
| 24 | Slc16a7 | 3982 | 0.240 | 0.2257 | Yes |
| 25 | Selenop | 4015 | 0.238 | 0.2347 | Yes |
| 26 | Prodh | 4022 | 0.238 | 0.2450 | Yes |
| 27 | Efhd1 | 4193 | 0.229 | 0.2466 | Yes |
| 28 | Idua | 4201 | 0.229 | 0.2564 | Yes |
| 29 | Zfp112 | 4599 | 0.209 | 0.2456 | Yes |
| 30 | Ypel1 | 4664 | 0.206 | 0.2515 | Yes |
| 31 | Thrb | 4904 | 0.193 | 0.2480 | Yes |
| 32 | Tnni3 | 5142 | 0.182 | 0.2440 | Yes |
| 33 | Fgfr3 | 5202 | 0.178 | 0.2490 | Yes |
| 34 | Dtnb | 5321 | 0.173 | 0.2507 | Yes |
| 35 | Ryr1 | 5393 | 0.169 | 0.2546 | Yes |
| 36 | Gamt | 5571 | 0.161 | 0.2528 | Yes |
| 37 | Tgfb2 | 5628 | 0.159 | 0.2571 | Yes |
| 38 | Clstn3 | 5792 | 0.152 | 0.2556 | Yes |
| 39 | Mefv | 5893 | 0.148 | 0.2571 | Yes |
| 40 | Celsr2 | 5916 | 0.147 | 0.2626 | Yes |
| 41 | Btg2 | 5970 | 0.144 | 0.2663 | Yes |
| 42 | Ryr2 | 6209 | 0.134 | 0.2602 | Yes |
| 43 | Thnsl2 | 6276 | 0.131 | 0.2626 | Yes |
| 44 | Hsd11b2 | 6331 | 0.129 | 0.2656 | Yes |
| 45 | Klk8 | 6407 | 0.126 | 0.2674 | Yes |
| 46 | Sptbn2 | 6474 | 0.123 | 0.2696 | Yes |
| 47 | Itgb1bp2 | 6669 | 0.114 | 0.2648 | No |
| 48 | Nos1 | 6725 | 0.114 | 0.2671 | No |
| 49 | Gtf3c5 | 7438 | 0.085 | 0.2346 | No |
| 50 | Prkn | 7483 | 0.083 | 0.2361 | No |
| 51 | Tfcp2l1 | 7797 | 0.072 | 0.2234 | No |
| 52 | Msh5 | 7862 | 0.069 | 0.2232 | No |
| 53 | Zc2hc1c | 7975 | 0.065 | 0.2204 | No |
| 54 | Mthfr | 8002 | 0.063 | 0.2219 | No |
| 55 | Stag3 | 8370 | 0.050 | 0.2055 | No |
| 56 | Ccdc106 | 8856 | 0.033 | 0.1823 | No |
| 57 | Pdk2 | 8874 | 0.032 | 0.1829 | No |
| 58 | Copz2 | 9002 | 0.028 | 0.1776 | No |
| 59 | Vps50 | 9206 | 0.020 | 0.1682 | No |
| 60 | Sgk1 | 9391 | 0.015 | 0.1595 | No |
| 61 | Asb7 | 9706 | 0.003 | 0.1437 | No |
| 62 | Capn9 | 9738 | 0.002 | 0.1422 | No |
| 63 | Tent5c | 9765 | 0.000 | 0.1409 | No |
| 64 | Tg | 9943 | -0.004 | 0.1321 | No |
| 65 | Kcnn1 | 10089 | -0.010 | 0.1251 | No |
| 66 | Cdkal1 | 10135 | -0.011 | 0.1234 | No |
| 67 | Bard1 | 10214 | -0.015 | 0.1200 | No |
| 68 | Skil | 10321 | -0.018 | 0.1155 | No |
| 69 | Abcg4 | 10637 | -0.029 | 0.1007 | No |
| 70 | Nr6a1 | 10788 | -0.035 | 0.0947 | No |
| 71 | Rgs11 | 10932 | -0.041 | 0.0892 | No |
| 72 | Myo15a | 10975 | -0.043 | 0.0890 | No |
| 73 | Synpo | 11098 | -0.047 | 0.0849 | No |
| 74 | Lfng | 11586 | -0.064 | 0.0630 | No |
| 75 | Slc5a5 | 11688 | -0.068 | 0.0609 | No |
| 76 | Tcf7l1 | 11736 | -0.070 | 0.0616 | No |
| 77 | Cpb1 | 11772 | -0.071 | 0.0630 | No |
| 78 | Edar | 11900 | -0.076 | 0.0600 | No |
| 79 | Snn | 13274 | -0.133 | -0.0039 | No |
| 80 | Rsad2 | 13384 | -0.137 | -0.0033 | No |
| 81 | Nrip2 | 13610 | -0.145 | -0.0083 | No |
| 82 | Grid2 | 13812 | -0.154 | -0.0116 | No |
| 83 | Pde6b | 14060 | -0.164 | -0.0169 | No |
| 84 | Cacna1f | 14085 | -0.165 | -0.0107 | No |
| 85 | Sidt1 | 14355 | -0.177 | -0.0165 | No |
| 86 | Atp4a | 14996 | -0.206 | -0.0399 | No |
| 87 | Ptprj | 15073 | -0.209 | -0.0344 | No |
| 88 | Bmpr1b | 15281 | -0.220 | -0.0351 | No |
| 89 | Dcc | 15415 | -0.227 | -0.0318 | No |
| 90 | Hc | 15618 | -0.238 | -0.0314 | No |
| 91 | Mfsd6 | 15643 | -0.239 | -0.0220 | No |
| 92 | Tex15 | 16625 | -0.294 | -0.0588 | No |
| 93 | Col2a1 | 16761 | -0.303 | -0.0521 | No |
| 94 | Arhgdig | 17333 | -0.348 | -0.0656 | No |
| 95 | Cyp39a1 | 17563 | -0.366 | -0.0610 | No |
| 96 | Entpd7 | 17579 | -0.367 | -0.0454 | No |
| 97 | Tgm1 | 17892 | -0.393 | -0.0437 | No |
| 98 | Cpeb3 | 18420 | -0.449 | -0.0505 | No |
| 99 | Kcnd1 | 18430 | -0.450 | -0.0309 | No |
| 100 | Cd80 | 18567 | -0.466 | -0.0170 | No |
| 101 | Tshb | 18720 | -0.486 | -0.0030 | No |
| 102 | Camk1d | 19326 | -0.616 | -0.0064 | No |
| 103 | Mx2 | 19419 | -0.649 | 0.0179 | No |