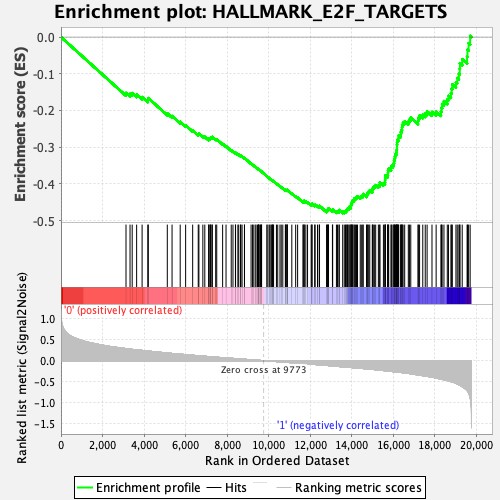
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_E2F_TARGETS |
| Enrichment Score (ES) | -0.47981134 |
| Normalized Enrichment Score (NES) | -1.9126239 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.005264847 |
| FWER p-Value | 0.008 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pold1 | 3131 | 0.287 | -0.1522 | No |
| 2 | Mlh1 | 3325 | 0.274 | -0.1547 | No |
| 3 | Zw10 | 3429 | 0.268 | -0.1527 | No |
| 4 | Wdr90 | 3643 | 0.257 | -0.1567 | No |
| 5 | Wee1 | 3906 | 0.244 | -0.1635 | No |
| 6 | Nbn | 4182 | 0.230 | -0.1714 | No |
| 7 | Exosc8 | 4204 | 0.229 | -0.1663 | No |
| 8 | Rad51c | 5120 | 0.183 | -0.2081 | No |
| 9 | Cenpm | 5349 | 0.171 | -0.2151 | No |
| 10 | Asf1b | 5741 | 0.154 | -0.2309 | No |
| 11 | Cks2 | 6003 | 0.143 | -0.2404 | No |
| 12 | Srsf2 | 6342 | 0.128 | -0.2542 | No |
| 13 | Cdkn3 | 6610 | 0.117 | -0.2647 | No |
| 14 | Depdc1a | 6642 | 0.115 | -0.2632 | No |
| 15 | Racgap1 | 6832 | 0.109 | -0.2699 | No |
| 16 | Pcna | 6926 | 0.105 | -0.2718 | No |
| 17 | Mcm4 | 7113 | 0.097 | -0.2787 | No |
| 18 | Eed | 7118 | 0.097 | -0.2763 | No |
| 19 | Pole | 7175 | 0.095 | -0.2766 | No |
| 20 | Cnot9 | 7194 | 0.095 | -0.2749 | No |
| 21 | Spag5 | 7220 | 0.094 | -0.2737 | No |
| 22 | Pan2 | 7280 | 0.091 | -0.2743 | No |
| 23 | Pold3 | 7285 | 0.091 | -0.2720 | No |
| 24 | Tubg1 | 7451 | 0.084 | -0.2782 | No |
| 25 | Dclre1b | 7501 | 0.082 | -0.2785 | No |
| 26 | Pola2 | 7785 | 0.072 | -0.2910 | No |
| 27 | Stmn1 | 7949 | 0.066 | -0.2976 | No |
| 28 | Hnrnpd | 8199 | 0.057 | -0.3087 | No |
| 29 | Myc | 8283 | 0.054 | -0.3115 | No |
| 30 | Ing3 | 8402 | 0.049 | -0.3162 | No |
| 31 | Cse1l | 8408 | 0.048 | -0.3152 | No |
| 32 | Pms2 | 8520 | 0.045 | -0.3197 | No |
| 33 | Ilf3 | 8561 | 0.044 | -0.3205 | No |
| 34 | Spc24 | 8648 | 0.041 | -0.3238 | No |
| 35 | Syncrip | 8661 | 0.040 | -0.3234 | No |
| 36 | Xrcc6 | 8721 | 0.038 | -0.3253 | No |
| 37 | Tipin | 8836 | 0.034 | -0.3303 | No |
| 38 | Rfc3 | 8839 | 0.034 | -0.3295 | No |
| 39 | Luc7l3 | 9155 | 0.023 | -0.3449 | No |
| 40 | Smc3 | 9227 | 0.020 | -0.3480 | No |
| 41 | Cdkn2a | 9256 | 0.019 | -0.3489 | No |
| 42 | Prkdc | 9348 | 0.016 | -0.3532 | No |
| 43 | Mre11a | 9394 | 0.014 | -0.3551 | No |
| 44 | Pds5b | 9465 | 0.011 | -0.3583 | No |
| 45 | Pole4 | 9510 | 0.010 | -0.3603 | No |
| 46 | Mcm2 | 9511 | 0.010 | -0.3601 | No |
| 47 | Stag1 | 9513 | 0.010 | -0.3599 | No |
| 48 | Ctcf | 9549 | 0.008 | -0.3614 | No |
| 49 | Mcm6 | 9601 | 0.006 | -0.3639 | No |
| 50 | Hus1 | 9617 | 0.005 | -0.3645 | No |
| 51 | Brms1l | 9657 | 0.004 | -0.3664 | No |
| 52 | Psmc3ip | 9662 | 0.004 | -0.3665 | No |
| 53 | Dut | 9912 | -0.003 | -0.3791 | No |
| 54 | Rad50 | 9964 | -0.005 | -0.3816 | No |
| 55 | Rfc1 | 10032 | -0.008 | -0.3848 | No |
| 56 | Mcm5 | 10075 | -0.009 | -0.3867 | No |
| 57 | Tra2b | 10146 | -0.012 | -0.3899 | No |
| 58 | Tubb5 | 10173 | -0.013 | -0.3909 | No |
| 59 | Rbbp7 | 10189 | -0.014 | -0.3913 | No |
| 60 | Bard1 | 10214 | -0.015 | -0.3921 | No |
| 61 | Shmt1 | 10239 | -0.016 | -0.3929 | No |
| 62 | Gspt1 | 10377 | -0.020 | -0.3994 | No |
| 63 | Msh2 | 10439 | -0.023 | -0.4019 | No |
| 64 | Dlgap5 | 10547 | -0.026 | -0.4067 | No |
| 65 | Atad2 | 10607 | -0.028 | -0.4089 | No |
| 66 | Mms22l | 10683 | -0.031 | -0.4119 | No |
| 67 | Gins1 | 10806 | -0.036 | -0.4172 | No |
| 68 | Suv39h1 | 10817 | -0.036 | -0.4167 | No |
| 69 | Hells | 10845 | -0.038 | -0.4171 | No |
| 70 | Chek2 | 10891 | -0.039 | -0.4183 | No |
| 71 | Rfc2 | 10894 | -0.040 | -0.4174 | No |
| 72 | Trp53 | 10896 | -0.040 | -0.4163 | No |
| 73 | Pold2 | 11117 | -0.048 | -0.4263 | No |
| 74 | Mybl2 | 11303 | -0.054 | -0.4343 | No |
| 75 | Nup107 | 11397 | -0.058 | -0.4375 | No |
| 76 | Rad1 | 11653 | -0.066 | -0.4487 | No |
| 77 | Mxd3 | 11699 | -0.068 | -0.4492 | No |
| 78 | Cdc20 | 11704 | -0.069 | -0.4475 | No |
| 79 | Ppp1r8 | 11711 | -0.069 | -0.4460 | No |
| 80 | Donson | 11793 | -0.072 | -0.4482 | No |
| 81 | Kif18b | 11875 | -0.075 | -0.4503 | No |
| 82 | Smc6 | 12049 | -0.083 | -0.4569 | No |
| 83 | Pttg1 | 12084 | -0.084 | -0.4564 | No |
| 84 | Ezh2 | 12090 | -0.084 | -0.4544 | No |
| 85 | Nolc1 | 12220 | -0.089 | -0.4585 | No |
| 86 | Pnn | 12238 | -0.090 | -0.4570 | No |
| 87 | Dek | 12355 | -0.094 | -0.4604 | No |
| 88 | Cit | 12436 | -0.097 | -0.4619 | No |
| 89 | Mcm7 | 12452 | -0.097 | -0.4600 | No |
| 90 | Nup153 | 12791 | -0.111 | -0.4743 | No |
| 91 | Ncapd2 | 12823 | -0.113 | -0.4728 | No |
| 92 | Hmgb2 | 12840 | -0.114 | -0.4706 | No |
| 93 | Mcm3 | 12878 | -0.115 | -0.4694 | No |
| 94 | Nudt21 | 12885 | -0.116 | -0.4665 | No |
| 95 | Rad21 | 13074 | -0.123 | -0.4728 | No |
| 96 | Ipo7 | 13085 | -0.124 | -0.4700 | No |
| 97 | Lmnb1 | 13266 | -0.132 | -0.4756 | No |
| 98 | Diaph3 | 13316 | -0.134 | -0.4745 | No |
| 99 | Eif2s1 | 13391 | -0.138 | -0.4746 | No |
| 100 | Asf1a | 13401 | -0.138 | -0.4713 | No |
| 101 | Dctpp1 | 13568 | -0.143 | -0.4759 | Yes |
| 102 | Gins4 | 13644 | -0.147 | -0.4758 | Yes |
| 103 | Plk1 | 13702 | -0.149 | -0.4747 | Yes |
| 104 | Slbp | 13736 | -0.151 | -0.4723 | Yes |
| 105 | Prim2 | 13774 | -0.152 | -0.4701 | Yes |
| 106 | Tmpo | 13802 | -0.154 | -0.4674 | Yes |
| 107 | Brca2 | 13867 | -0.156 | -0.4664 | Yes |
| 108 | Cdc25b | 13879 | -0.157 | -0.4627 | Yes |
| 109 | Ube2s | 13961 | -0.160 | -0.4626 | Yes |
| 110 | Ubr7 | 13968 | -0.160 | -0.4586 | Yes |
| 111 | Timeless | 13974 | -0.161 | -0.4545 | Yes |
| 112 | Dnmt1 | 14010 | -0.162 | -0.4519 | Yes |
| 113 | H2ax | 14021 | -0.163 | -0.4480 | Yes |
| 114 | E2f8 | 14048 | -0.164 | -0.4449 | Yes |
| 115 | Rpa1 | 14112 | -0.167 | -0.4436 | Yes |
| 116 | Lig1 | 14126 | -0.167 | -0.4398 | Yes |
| 117 | Nap1l1 | 14189 | -0.169 | -0.4384 | Yes |
| 118 | Cks1b | 14232 | -0.171 | -0.4359 | Yes |
| 119 | Orc6 | 14282 | -0.174 | -0.4337 | Yes |
| 120 | Nup205 | 14413 | -0.179 | -0.4355 | Yes |
| 121 | Xpo1 | 14465 | -0.182 | -0.4333 | Yes |
| 122 | Snrpb | 14535 | -0.185 | -0.4318 | Yes |
| 123 | Pop7 | 14554 | -0.186 | -0.4277 | Yes |
| 124 | Tbrg4 | 14716 | -0.192 | -0.4308 | Yes |
| 125 | Trip13 | 14749 | -0.194 | -0.4272 | Yes |
| 126 | Usp1 | 14777 | -0.195 | -0.4233 | Yes |
| 127 | Ak2 | 14823 | -0.197 | -0.4203 | Yes |
| 128 | Lsm8 | 14872 | -0.199 | -0.4174 | Yes |
| 129 | Nop56 | 14995 | -0.206 | -0.4181 | Yes |
| 130 | Cdkn1a | 15001 | -0.206 | -0.4128 | Yes |
| 131 | Ung | 15040 | -0.208 | -0.4091 | Yes |
| 132 | Smc4 | 15093 | -0.210 | -0.4061 | Yes |
| 133 | Cenpe | 15157 | -0.213 | -0.4036 | Yes |
| 134 | Psip1 | 15287 | -0.220 | -0.4042 | Yes |
| 135 | Mad2l1 | 15352 | -0.223 | -0.4015 | Yes |
| 136 | Ranbp1 | 15359 | -0.224 | -0.3958 | Yes |
| 137 | H2az1 | 15529 | -0.233 | -0.3981 | Yes |
| 138 | Pa2g4 | 15603 | -0.237 | -0.3954 | Yes |
| 139 | Cdca3 | 15610 | -0.238 | -0.3893 | Yes |
| 140 | Ppm1d | 15616 | -0.238 | -0.3832 | Yes |
| 141 | Kif2c | 15619 | -0.238 | -0.3769 | Yes |
| 142 | Cdkn1b | 15732 | -0.244 | -0.3760 | Yes |
| 143 | Rpa2 | 15747 | -0.245 | -0.3701 | Yes |
| 144 | Jpt1 | 15753 | -0.245 | -0.3638 | Yes |
| 145 | Dscc1 | 15783 | -0.247 | -0.3586 | Yes |
| 146 | Ddx39a | 15897 | -0.253 | -0.3575 | Yes |
| 147 | Mki67 | 15910 | -0.254 | -0.3513 | Yes |
| 148 | Prps1 | 15988 | -0.258 | -0.3483 | Yes |
| 149 | Nasp | 16031 | -0.260 | -0.3434 | Yes |
| 150 | Orc2 | 16043 | -0.261 | -0.3369 | Yes |
| 151 | Cbx5 | 16064 | -0.262 | -0.3309 | Yes |
| 152 | Chek1 | 16086 | -0.264 | -0.3248 | Yes |
| 153 | Cdc25a | 16110 | -0.265 | -0.3189 | Yes |
| 154 | Plk4 | 16157 | -0.267 | -0.3140 | Yes |
| 155 | Ccnb2 | 16170 | -0.267 | -0.3074 | Yes |
| 156 | Smc1a | 16180 | -0.268 | -0.3007 | Yes |
| 157 | Espl1 | 16181 | -0.268 | -0.2935 | Yes |
| 158 | Kif22 | 16190 | -0.268 | -0.2866 | Yes |
| 159 | Phf5a | 16203 | -0.269 | -0.2800 | Yes |
| 160 | Brca1 | 16260 | -0.273 | -0.2755 | Yes |
| 161 | Tcf19 | 16263 | -0.273 | -0.2683 | Yes |
| 162 | Lyar | 16361 | -0.278 | -0.2657 | Yes |
| 163 | Bub1b | 16370 | -0.279 | -0.2586 | Yes |
| 164 | Ccp110 | 16405 | -0.281 | -0.2528 | Yes |
| 165 | Anp32e | 16426 | -0.282 | -0.2462 | Yes |
| 166 | Cdk4 | 16434 | -0.282 | -0.2389 | Yes |
| 167 | Tk1 | 16476 | -0.285 | -0.2334 | Yes |
| 168 | Lbr | 16554 | -0.290 | -0.2295 | Yes |
| 169 | Paics | 16729 | -0.301 | -0.2302 | Yes |
| 170 | Cdca8 | 16782 | -0.305 | -0.2247 | Yes |
| 171 | Hmgb3 | 16847 | -0.309 | -0.2196 | Yes |
| 172 | Top2a | 17196 | -0.338 | -0.2283 | Yes |
| 173 | Ube2t | 17211 | -0.340 | -0.2198 | Yes |
| 174 | Melk | 17271 | -0.343 | -0.2136 | Yes |
| 175 | Kif4 | 17425 | -0.354 | -0.2119 | Yes |
| 176 | Rad51ap1 | 17544 | -0.365 | -0.2081 | Yes |
| 177 | Aurkb | 17634 | -0.371 | -0.2026 | Yes |
| 178 | Hmga1b | 17869 | -0.391 | -0.2040 | Yes |
| 179 | Rnaseh2a | 18069 | -0.409 | -0.2032 | Yes |
| 180 | Srsf1 | 18300 | -0.435 | -0.2032 | Yes |
| 181 | Mthfd2 | 18325 | -0.438 | -0.1926 | Yes |
| 182 | Dck | 18358 | -0.441 | -0.1823 | Yes |
| 183 | Gins3 | 18445 | -0.452 | -0.1745 | Yes |
| 184 | Rrm2 | 18606 | -0.470 | -0.1701 | Yes |
| 185 | Rpa3 | 18662 | -0.477 | -0.1600 | Yes |
| 186 | Tacc3 | 18781 | -0.496 | -0.1527 | Yes |
| 187 | Cdkn2c | 18816 | -0.501 | -0.1409 | Yes |
| 188 | Hmmr | 18847 | -0.505 | -0.1288 | Yes |
| 189 | Cdk1 | 19023 | -0.536 | -0.1233 | Yes |
| 190 | Ssrp1 | 19085 | -0.552 | -0.1115 | Yes |
| 191 | Spc25 | 19165 | -0.571 | -0.1002 | Yes |
| 192 | Tfrc | 19206 | -0.580 | -0.0866 | Yes |
| 193 | Ran | 19214 | -0.584 | -0.0712 | Yes |
| 194 | Nme1 | 19325 | -0.615 | -0.0602 | Yes |
| 195 | Birc5 | 19560 | -0.708 | -0.0531 | Yes |
| 196 | Aurka | 19579 | -0.719 | -0.0346 | Yes |
| 197 | Ccne1 | 19632 | -0.763 | -0.0167 | Yes |
| 198 | Prdx4 | 19707 | -0.882 | 0.0033 | Yes |