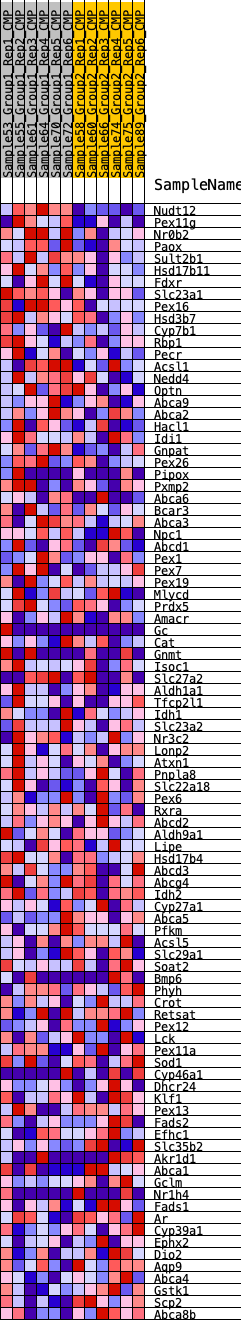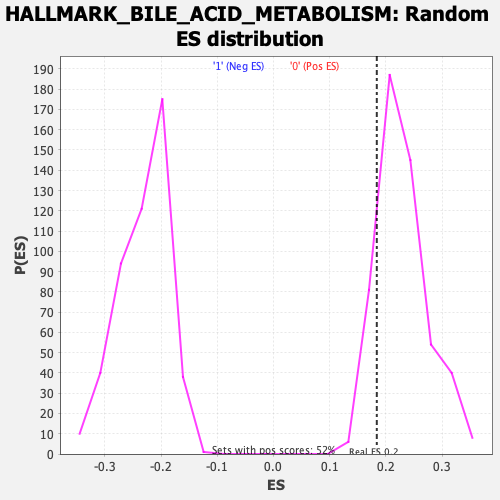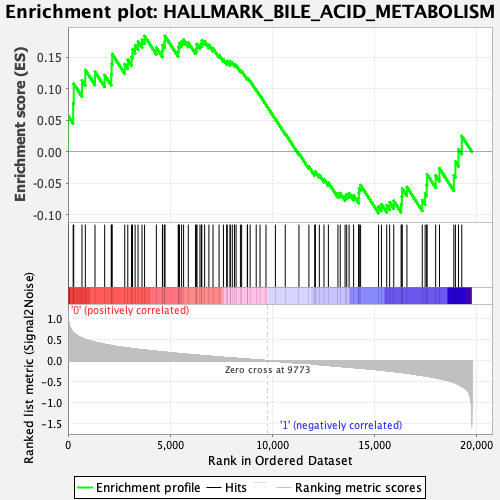
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.18390441 |
| Normalized Enrichment Score (NES) | 0.80560696 |
| Nominal p-value | 0.8502879 |
| FDR q-value | 0.9977779 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nudt12 | 1 | 1.183 | 0.0574 | Yes |
| 2 | Pex11g | 253 | 0.670 | 0.0772 | Yes |
| 3 | Nr0b2 | 280 | 0.655 | 0.1077 | Yes |
| 4 | Paox | 684 | 0.538 | 0.1134 | Yes |
| 5 | Sult2b1 | 848 | 0.507 | 0.1297 | Yes |
| 6 | Hsd17b11 | 1322 | 0.437 | 0.1269 | Yes |
| 7 | Fdxr | 1795 | 0.387 | 0.1218 | Yes |
| 8 | Slc23a1 | 2116 | 0.358 | 0.1229 | Yes |
| 9 | Pex16 | 2135 | 0.357 | 0.1393 | Yes |
| 10 | Hsd3b7 | 2163 | 0.354 | 0.1552 | Yes |
| 11 | Cyp7b1 | 2778 | 0.309 | 0.1390 | Yes |
| 12 | Rbp1 | 2927 | 0.300 | 0.1461 | Yes |
| 13 | Pecr | 3110 | 0.288 | 0.1508 | Yes |
| 14 | Acsl1 | 3158 | 0.286 | 0.1623 | Yes |
| 15 | Nedd4 | 3286 | 0.277 | 0.1693 | Yes |
| 16 | Optn | 3430 | 0.268 | 0.1751 | Yes |
| 17 | Abca9 | 3624 | 0.258 | 0.1778 | Yes |
| 18 | Abca2 | 3749 | 0.251 | 0.1837 | Yes |
| 19 | Hacl1 | 4326 | 0.222 | 0.1652 | Yes |
| 20 | Idi1 | 4623 | 0.208 | 0.1603 | Yes |
| 21 | Gnpat | 4639 | 0.207 | 0.1696 | Yes |
| 22 | Pex26 | 4737 | 0.202 | 0.1744 | Yes |
| 23 | Pipox | 4744 | 0.201 | 0.1839 | Yes |
| 24 | Pxmp2 | 5396 | 0.169 | 0.1590 | No |
| 25 | Abca6 | 5415 | 0.168 | 0.1663 | No |
| 26 | Bcar3 | 5457 | 0.167 | 0.1723 | No |
| 27 | Abca3 | 5562 | 0.162 | 0.1749 | No |
| 28 | Npc1 | 5660 | 0.158 | 0.1777 | No |
| 29 | Abcd1 | 5890 | 0.148 | 0.1733 | No |
| 30 | Pex1 | 6241 | 0.132 | 0.1619 | No |
| 31 | Pex7 | 6307 | 0.130 | 0.1649 | No |
| 32 | Pex19 | 6308 | 0.130 | 0.1712 | No |
| 33 | Mlycd | 6461 | 0.124 | 0.1695 | No |
| 34 | Prdx5 | 6529 | 0.120 | 0.1719 | No |
| 35 | Amacr | 6553 | 0.120 | 0.1766 | No |
| 36 | Gc | 6690 | 0.114 | 0.1752 | No |
| 37 | Cat | 6899 | 0.106 | 0.1697 | No |
| 38 | Gnmt | 7100 | 0.098 | 0.1643 | No |
| 39 | Isoc1 | 7396 | 0.087 | 0.1536 | No |
| 40 | Slc27a2 | 7610 | 0.077 | 0.1465 | No |
| 41 | Aldh1a1 | 7769 | 0.073 | 0.1420 | No |
| 42 | Tfcp2l1 | 7797 | 0.072 | 0.1441 | No |
| 43 | Idh1 | 7936 | 0.066 | 0.1403 | No |
| 44 | Slc23a2 | 7942 | 0.066 | 0.1433 | No |
| 45 | Nr3c2 | 8044 | 0.062 | 0.1411 | No |
| 46 | Lonp2 | 8149 | 0.059 | 0.1387 | No |
| 47 | Atxn1 | 8228 | 0.055 | 0.1374 | No |
| 48 | Pnpla8 | 8442 | 0.047 | 0.1289 | No |
| 49 | Slc22a18 | 8502 | 0.045 | 0.1281 | No |
| 50 | Pex6 | 8778 | 0.036 | 0.1159 | No |
| 51 | Rxra | 8790 | 0.036 | 0.1171 | No |
| 52 | Abcd2 | 8917 | 0.031 | 0.1122 | No |
| 53 | Aldh9a1 | 9215 | 0.020 | 0.0980 | No |
| 54 | Lipe | 9401 | 0.014 | 0.0893 | No |
| 55 | Hsd17b4 | 9693 | 0.003 | 0.0747 | No |
| 56 | Abcd3 | 10154 | -0.012 | 0.0519 | No |
| 57 | Abcg4 | 10637 | -0.029 | 0.0288 | No |
| 58 | Idh2 | 11307 | -0.054 | -0.0025 | No |
| 59 | Cyp27a1 | 11795 | -0.072 | -0.0238 | No |
| 60 | Abca5 | 12078 | -0.084 | -0.0340 | No |
| 61 | Pfkm | 12113 | -0.085 | -0.0316 | No |
| 62 | Acsl5 | 12305 | -0.092 | -0.0369 | No |
| 63 | Slc29a1 | 12534 | -0.100 | -0.0436 | No |
| 64 | Soat2 | 12749 | -0.109 | -0.0492 | No |
| 65 | Bmp6 | 13217 | -0.130 | -0.0666 | No |
| 66 | Phyh | 13329 | -0.135 | -0.0656 | No |
| 67 | Crot | 13566 | -0.143 | -0.0707 | No |
| 68 | Retsat | 13645 | -0.147 | -0.0675 | No |
| 69 | Pex12 | 13763 | -0.152 | -0.0661 | No |
| 70 | Lck | 13983 | -0.161 | -0.0694 | No |
| 71 | Pex11a | 14235 | -0.172 | -0.0738 | No |
| 72 | Sod1 | 14241 | -0.172 | -0.0657 | No |
| 73 | Cyp46a1 | 14253 | -0.172 | -0.0579 | No |
| 74 | Dhcr24 | 14326 | -0.176 | -0.0530 | No |
| 75 | Klf1 | 15205 | -0.216 | -0.0872 | No |
| 76 | Pex13 | 15345 | -0.223 | -0.0834 | No |
| 77 | Fads2 | 15605 | -0.237 | -0.0850 | No |
| 78 | Efhc1 | 15748 | -0.245 | -0.0804 | No |
| 79 | Slc35b2 | 15949 | -0.256 | -0.0781 | No |
| 80 | Akr1d1 | 16310 | -0.276 | -0.0830 | No |
| 81 | Abca1 | 16346 | -0.277 | -0.0713 | No |
| 82 | Gclm | 16360 | -0.278 | -0.0584 | No |
| 83 | Nr1h4 | 16594 | -0.293 | -0.0560 | No |
| 84 | Fads1 | 17345 | -0.348 | -0.0772 | No |
| 85 | Ar | 17486 | -0.360 | -0.0668 | No |
| 86 | Cyp39a1 | 17563 | -0.366 | -0.0529 | No |
| 87 | Ephx2 | 17581 | -0.367 | -0.0360 | No |
| 88 | Dio2 | 18001 | -0.402 | -0.0377 | No |
| 89 | Aqp9 | 18187 | -0.423 | -0.0265 | No |
| 90 | Abca4 | 18891 | -0.512 | -0.0374 | No |
| 91 | Gstk1 | 18968 | -0.525 | -0.0157 | No |
| 92 | Scp2 | 19119 | -0.561 | 0.0039 | No |
| 93 | Abca8b | 19279 | -0.601 | 0.0250 | No |