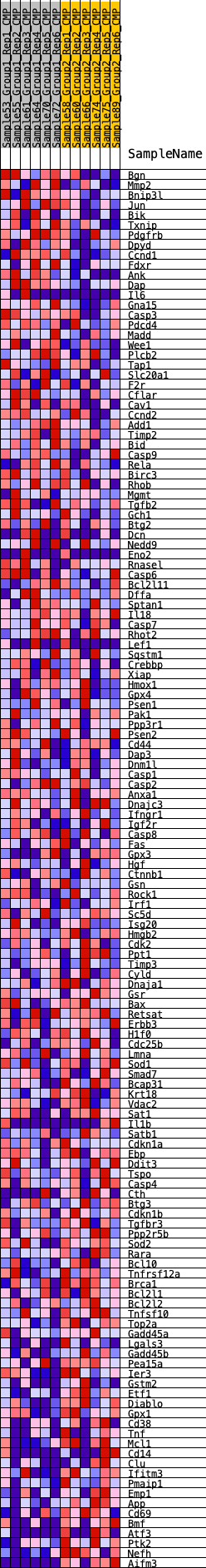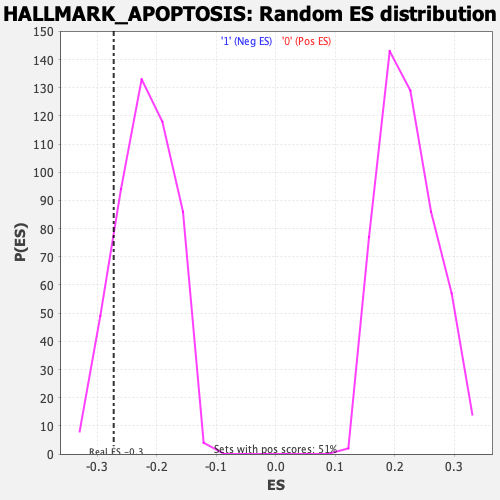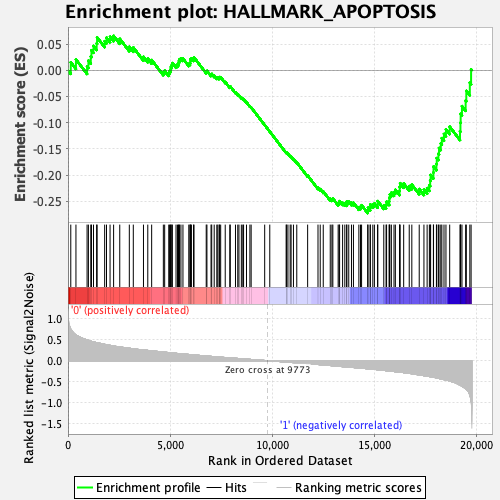
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CMP.Basophil_Pheno.cls #Group1_versus_Group2.Basophil_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | Basophil_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_APOPTOSIS |
| Enrichment Score (ES) | -0.27262205 |
| Normalized Enrichment Score (NES) | -1.2387744 |
| Nominal p-value | 0.15243903 |
| FDR q-value | 0.31315497 |
| FWER p-Value | 0.861 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Bgn | 136 | 0.747 | 0.0152 | No |
| 2 | Mmp2 | 389 | 0.613 | 0.0206 | No |
| 3 | Bnip3l | 934 | 0.493 | 0.0075 | No |
| 4 | Jun | 999 | 0.481 | 0.0185 | No |
| 5 | Bik | 1116 | 0.463 | 0.0263 | No |
| 6 | Txnip | 1149 | 0.458 | 0.0382 | No |
| 7 | Pdgfrb | 1248 | 0.445 | 0.0464 | No |
| 8 | Dpyd | 1403 | 0.427 | 0.0513 | No |
| 9 | Ccnd1 | 1424 | 0.426 | 0.0629 | No |
| 10 | Fdxr | 1795 | 0.387 | 0.0555 | No |
| 11 | Ank | 1885 | 0.379 | 0.0622 | No |
| 12 | Dap | 2059 | 0.363 | 0.0642 | No |
| 13 | Il6 | 2233 | 0.348 | 0.0657 | No |
| 14 | Gna15 | 2534 | 0.326 | 0.0600 | No |
| 15 | Casp3 | 3002 | 0.296 | 0.0450 | No |
| 16 | Pdcd4 | 3197 | 0.283 | 0.0435 | No |
| 17 | Madd | 3695 | 0.254 | 0.0257 | No |
| 18 | Wee1 | 3906 | 0.244 | 0.0223 | No |
| 19 | Plcb2 | 4096 | 0.234 | 0.0196 | No |
| 20 | Tap1 | 4668 | 0.205 | -0.0034 | No |
| 21 | Slc20a1 | 4733 | 0.202 | -0.0007 | No |
| 22 | F2r | 4933 | 0.192 | -0.0051 | No |
| 23 | Cflar | 4973 | 0.189 | -0.0015 | No |
| 24 | Cav1 | 5020 | 0.188 | 0.0017 | No |
| 25 | Ccnd2 | 5039 | 0.187 | 0.0063 | No |
| 26 | Add1 | 5080 | 0.185 | 0.0098 | No |
| 27 | Timp2 | 5109 | 0.184 | 0.0138 | No |
| 28 | Bid | 5289 | 0.175 | 0.0098 | No |
| 29 | Casp9 | 5360 | 0.171 | 0.0113 | No |
| 30 | Rela | 5410 | 0.169 | 0.0138 | No |
| 31 | Birc3 | 5432 | 0.168 | 0.0177 | No |
| 32 | Rhob | 5462 | 0.167 | 0.0212 | No |
| 33 | Mgmt | 5529 | 0.164 | 0.0227 | No |
| 34 | Tgfb2 | 5628 | 0.159 | 0.0224 | No |
| 35 | Gch1 | 5914 | 0.148 | 0.0123 | No |
| 36 | Btg2 | 5970 | 0.144 | 0.0138 | No |
| 37 | Dcn | 5991 | 0.143 | 0.0170 | No |
| 38 | Nedd9 | 5992 | 0.143 | 0.0212 | No |
| 39 | Eno2 | 6043 | 0.140 | 0.0228 | No |
| 40 | Rnasel | 6146 | 0.136 | 0.0217 | No |
| 41 | Casp6 | 6170 | 0.135 | 0.0245 | No |
| 42 | Bcl2l11 | 6767 | 0.112 | -0.0025 | No |
| 43 | Dffa | 6802 | 0.110 | -0.0010 | No |
| 44 | Sptan1 | 7003 | 0.101 | -0.0082 | No |
| 45 | Il18 | 7037 | 0.100 | -0.0069 | No |
| 46 | Casp7 | 7158 | 0.096 | -0.0101 | No |
| 47 | Rhot2 | 7286 | 0.090 | -0.0139 | No |
| 48 | Lef1 | 7334 | 0.089 | -0.0137 | No |
| 49 | Sqstm1 | 7401 | 0.086 | -0.0145 | No |
| 50 | Crebbp | 7421 | 0.086 | -0.0129 | No |
| 51 | Xiap | 7475 | 0.083 | -0.0132 | No |
| 52 | Hmox1 | 7698 | 0.075 | -0.0223 | No |
| 53 | Gpx4 | 7922 | 0.067 | -0.0316 | No |
| 54 | Psen1 | 7944 | 0.066 | -0.0308 | No |
| 55 | Pak1 | 8200 | 0.057 | -0.0421 | No |
| 56 | Ppp3r1 | 8312 | 0.053 | -0.0462 | No |
| 57 | Psen2 | 8369 | 0.050 | -0.0475 | No |
| 58 | Cd44 | 8487 | 0.046 | -0.0521 | No |
| 59 | Dap3 | 8559 | 0.044 | -0.0544 | No |
| 60 | Dnm1l | 8596 | 0.042 | -0.0550 | No |
| 61 | Casp1 | 8739 | 0.038 | -0.0611 | No |
| 62 | Casp2 | 8906 | 0.031 | -0.0687 | No |
| 63 | Anxa1 | 8967 | 0.029 | -0.0709 | No |
| 64 | Dnajc3 | 9630 | 0.005 | -0.1044 | No |
| 65 | Ifngr1 | 9879 | -0.002 | -0.1170 | No |
| 66 | Igf2r | 10680 | -0.031 | -0.1568 | No |
| 67 | Casp8 | 10703 | -0.032 | -0.1570 | No |
| 68 | Fas | 10768 | -0.034 | -0.1593 | No |
| 69 | Gpx3 | 10877 | -0.039 | -0.1636 | No |
| 70 | Hgf | 10923 | -0.041 | -0.1647 | No |
| 71 | Ctnnb1 | 11038 | -0.045 | -0.1692 | No |
| 72 | Gsn | 11201 | -0.051 | -0.1759 | No |
| 73 | Rock1 | 11733 | -0.070 | -0.2009 | No |
| 74 | Irf1 | 12240 | -0.090 | -0.2240 | No |
| 75 | Sc5d | 12349 | -0.094 | -0.2267 | No |
| 76 | Isg20 | 12497 | -0.099 | -0.2313 | No |
| 77 | Hmgb2 | 12840 | -0.114 | -0.2453 | No |
| 78 | Cdk2 | 12921 | -0.117 | -0.2459 | No |
| 79 | Ppt1 | 12967 | -0.118 | -0.2447 | No |
| 80 | Timp3 | 13227 | -0.131 | -0.2540 | No |
| 81 | Cyld | 13276 | -0.133 | -0.2525 | No |
| 82 | Dnaja1 | 13298 | -0.134 | -0.2497 | No |
| 83 | Gsr | 13445 | -0.140 | -0.2530 | No |
| 84 | Bax | 13553 | -0.143 | -0.2542 | No |
| 85 | Retsat | 13645 | -0.147 | -0.2545 | No |
| 86 | Erbb3 | 13650 | -0.147 | -0.2503 | No |
| 87 | H1f0 | 13746 | -0.151 | -0.2507 | No |
| 88 | Cdc25b | 13879 | -0.157 | -0.2528 | No |
| 89 | Lmna | 13975 | -0.161 | -0.2528 | No |
| 90 | Sod1 | 14241 | -0.172 | -0.2612 | No |
| 91 | Smad7 | 14322 | -0.176 | -0.2601 | No |
| 92 | Bcap31 | 14374 | -0.178 | -0.2574 | No |
| 93 | Krt18 | 14673 | -0.190 | -0.2670 | Yes |
| 94 | Vdac2 | 14689 | -0.191 | -0.2621 | Yes |
| 95 | Sat1 | 14796 | -0.196 | -0.2617 | Yes |
| 96 | Il1b | 14800 | -0.196 | -0.2560 | Yes |
| 97 | Satb1 | 14924 | -0.202 | -0.2563 | Yes |
| 98 | Cdkn1a | 15001 | -0.206 | -0.2541 | Yes |
| 99 | Ebp | 15162 | -0.213 | -0.2559 | Yes |
| 100 | Ddit3 | 15163 | -0.214 | -0.2496 | Yes |
| 101 | Tspo | 15461 | -0.229 | -0.2579 | Yes |
| 102 | Casp4 | 15573 | -0.235 | -0.2566 | Yes |
| 103 | Cth | 15598 | -0.237 | -0.2508 | Yes |
| 104 | Btg3 | 15727 | -0.244 | -0.2501 | Yes |
| 105 | Cdkn1b | 15732 | -0.244 | -0.2431 | Yes |
| 106 | Tgfbr3 | 15758 | -0.246 | -0.2370 | Yes |
| 107 | Ppp2r5b | 15832 | -0.250 | -0.2334 | Yes |
| 108 | Sod2 | 15961 | -0.256 | -0.2323 | Yes |
| 109 | Rara | 16028 | -0.260 | -0.2279 | Yes |
| 110 | Bcl10 | 16233 | -0.270 | -0.2303 | Yes |
| 111 | Tnfrsf12a | 16239 | -0.271 | -0.2225 | Yes |
| 112 | Brca1 | 16260 | -0.273 | -0.2155 | Yes |
| 113 | Bcl2l1 | 16435 | -0.282 | -0.2159 | Yes |
| 114 | Bcl2l2 | 16707 | -0.298 | -0.2209 | Yes |
| 115 | Tnfsf10 | 16834 | -0.309 | -0.2182 | Yes |
| 116 | Top2a | 17196 | -0.338 | -0.2266 | Yes |
| 117 | Gadd45a | 17427 | -0.354 | -0.2278 | Yes |
| 118 | Lgals3 | 17583 | -0.367 | -0.2248 | Yes |
| 119 | Gadd45b | 17693 | -0.375 | -0.2192 | Yes |
| 120 | Pea15a | 17732 | -0.378 | -0.2100 | Yes |
| 121 | Ier3 | 17754 | -0.380 | -0.1998 | Yes |
| 122 | Gstm2 | 17890 | -0.393 | -0.1950 | Yes |
| 123 | Etf1 | 17897 | -0.394 | -0.1836 | Yes |
| 124 | Diablo | 18038 | -0.406 | -0.1787 | Yes |
| 125 | Gpx1 | 18055 | -0.408 | -0.1674 | Yes |
| 126 | Cd38 | 18135 | -0.417 | -0.1591 | Yes |
| 127 | Tnf | 18166 | -0.421 | -0.1482 | Yes |
| 128 | Mcl1 | 18252 | -0.431 | -0.1397 | Yes |
| 129 | Cd14 | 18304 | -0.436 | -0.1294 | Yes |
| 130 | Clu | 18409 | -0.448 | -0.1214 | Yes |
| 131 | Ifitm3 | 18505 | -0.459 | -0.1126 | Yes |
| 132 | Pmaip1 | 18688 | -0.481 | -0.1076 | Yes |
| 133 | Emp1 | 19193 | -0.577 | -0.1162 | Yes |
| 134 | App | 19215 | -0.584 | -0.1000 | Yes |
| 135 | Cd69 | 19226 | -0.587 | -0.0831 | Yes |
| 136 | Bmf | 19289 | -0.603 | -0.0684 | Yes |
| 137 | Atf3 | 19469 | -0.665 | -0.0578 | Yes |
| 138 | Ptk2 | 19502 | -0.679 | -0.0393 | Yes |
| 139 | Nefh | 19674 | -0.820 | -0.0237 | Yes |
| 140 | Aifm3 | 19736 | -0.964 | 0.0018 | Yes |