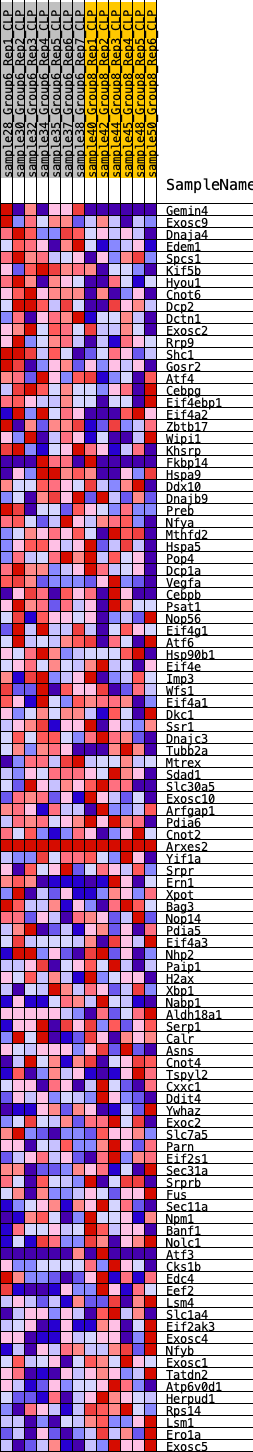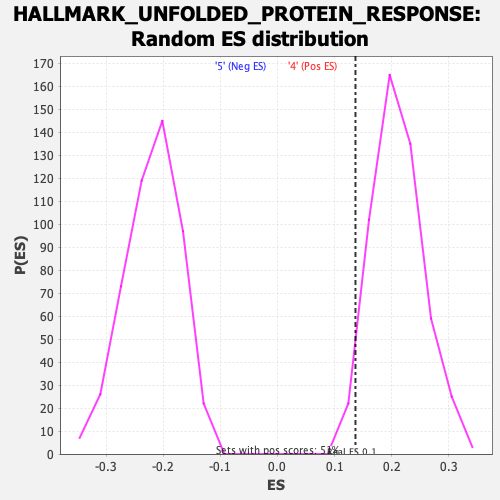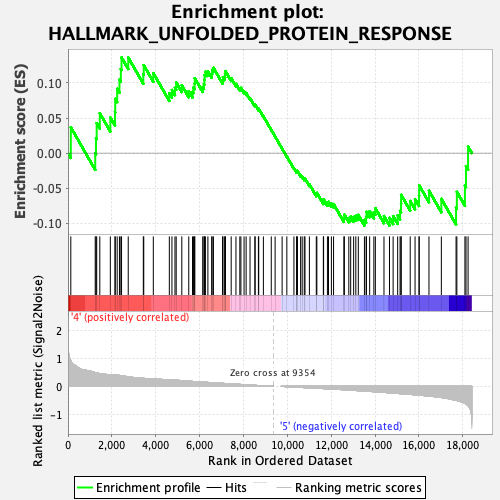
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.13645492 |
| Normalized Enrichment Score (NES) | 0.6487723 |
| Nominal p-value | 0.9765166 |
| FDR q-value | 0.96516937 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gemin4 | 128 | 0.913 | 0.0368 | Yes |
| 2 | Exosc9 | 1241 | 0.493 | -0.0002 | Yes |
| 3 | Dnaja4 | 1276 | 0.488 | 0.0213 | Yes |
| 4 | Edem1 | 1308 | 0.480 | 0.0427 | Yes |
| 5 | Spcs1 | 1450 | 0.457 | 0.0569 | Yes |
| 6 | Kif5b | 1930 | 0.416 | 0.0507 | Yes |
| 7 | Hyou1 | 2142 | 0.405 | 0.0586 | Yes |
| 8 | Cnot6 | 2152 | 0.403 | 0.0775 | Yes |
| 9 | Dcp2 | 2242 | 0.399 | 0.0917 | Yes |
| 10 | Dctn1 | 2346 | 0.391 | 0.1049 | Yes |
| 11 | Exosc2 | 2409 | 0.383 | 0.1199 | Yes |
| 12 | Rrp9 | 2441 | 0.380 | 0.1365 | Yes |
| 13 | Shc1 | 2746 | 0.344 | 0.1364 | No |
| 14 | Gosr2 | 3433 | 0.288 | 0.1127 | No |
| 15 | Atf4 | 3453 | 0.286 | 0.1254 | No |
| 16 | Cebpg | 3889 | 0.261 | 0.1141 | No |
| 17 | Eif4ebp1 | 4619 | 0.235 | 0.0856 | No |
| 18 | Eif4a2 | 4738 | 0.227 | 0.0900 | No |
| 19 | Zbtb17 | 4876 | 0.224 | 0.0933 | No |
| 20 | Wipi1 | 4932 | 0.220 | 0.1009 | No |
| 21 | Khsrp | 5188 | 0.204 | 0.0967 | No |
| 22 | Fkbp14 | 5505 | 0.186 | 0.0884 | No |
| 23 | Hspa9 | 5676 | 0.176 | 0.0875 | No |
| 24 | Ddx10 | 5719 | 0.174 | 0.0936 | No |
| 25 | Dnajb9 | 5771 | 0.171 | 0.0990 | No |
| 26 | Preb | 5779 | 0.170 | 0.1068 | No |
| 27 | Nfya | 6140 | 0.152 | 0.0944 | No |
| 28 | Mthfd2 | 6197 | 0.149 | 0.0985 | No |
| 29 | Hspa5 | 6216 | 0.148 | 0.1046 | No |
| 30 | Pop4 | 6225 | 0.147 | 0.1112 | No |
| 31 | Dcp1a | 6264 | 0.145 | 0.1161 | No |
| 32 | Vegfa | 6377 | 0.140 | 0.1167 | No |
| 33 | Cebpb | 6551 | 0.131 | 0.1135 | No |
| 34 | Psat1 | 6569 | 0.130 | 0.1188 | No |
| 35 | Nop56 | 6630 | 0.127 | 0.1217 | No |
| 36 | Eif4g1 | 7045 | 0.108 | 0.1042 | No |
| 37 | Atf6 | 7065 | 0.107 | 0.1083 | No |
| 38 | Hsp90b1 | 7142 | 0.103 | 0.1091 | No |
| 39 | Eif4e | 7161 | 0.101 | 0.1129 | No |
| 40 | Imp3 | 7177 | 0.100 | 0.1169 | No |
| 41 | Wfs1 | 7439 | 0.088 | 0.1069 | No |
| 42 | Eif4a1 | 7656 | 0.077 | 0.0988 | No |
| 43 | Dkc1 | 7833 | 0.070 | 0.0926 | No |
| 44 | Ssr1 | 7882 | 0.067 | 0.0932 | No |
| 45 | Dnajc3 | 8026 | 0.059 | 0.0882 | No |
| 46 | Tubb2a | 8120 | 0.055 | 0.0858 | No |
| 47 | Mtrex | 8298 | 0.047 | 0.0784 | No |
| 48 | Sdad1 | 8522 | 0.036 | 0.0679 | No |
| 49 | Slc30a5 | 8527 | 0.036 | 0.0694 | No |
| 50 | Exosc10 | 8680 | 0.029 | 0.0625 | No |
| 51 | Arfgap1 | 8694 | 0.029 | 0.0631 | No |
| 52 | Pdia6 | 8907 | 0.019 | 0.0525 | No |
| 53 | Cnot2 | 9266 | 0.004 | 0.0331 | No |
| 54 | Arxes2 | 9441 | 0.000 | 0.0236 | No |
| 55 | Yif1a | 9761 | -0.002 | 0.0063 | No |
| 56 | Srpr | 9976 | -0.011 | -0.0049 | No |
| 57 | Ern1 | 10299 | -0.024 | -0.0213 | No |
| 58 | Xpot | 10421 | -0.030 | -0.0265 | No |
| 59 | Bag3 | 10429 | -0.030 | -0.0254 | No |
| 60 | Nop14 | 10459 | -0.031 | -0.0255 | No |
| 61 | Pdia5 | 10604 | -0.038 | -0.0316 | No |
| 62 | Eif4a3 | 10675 | -0.039 | -0.0335 | No |
| 63 | Nhp2 | 10770 | -0.044 | -0.0365 | No |
| 64 | Paip1 | 10803 | -0.045 | -0.0361 | No |
| 65 | H2ax | 11004 | -0.054 | -0.0444 | No |
| 66 | Xbp1 | 11318 | -0.065 | -0.0584 | No |
| 67 | Nabp1 | 11344 | -0.066 | -0.0566 | No |
| 68 | Aldh18a1 | 11640 | -0.079 | -0.0690 | No |
| 69 | Serp1 | 11652 | -0.079 | -0.0658 | No |
| 70 | Calr | 11826 | -0.088 | -0.0710 | No |
| 71 | Asns | 11875 | -0.088 | -0.0694 | No |
| 72 | Cnot4 | 12007 | -0.095 | -0.0720 | No |
| 73 | Tspyl2 | 12106 | -0.099 | -0.0726 | No |
| 74 | Cxxc1 | 12576 | -0.121 | -0.0924 | No |
| 75 | Ddit4 | 12598 | -0.122 | -0.0877 | No |
| 76 | Ywhaz | 12801 | -0.131 | -0.0924 | No |
| 77 | Exoc2 | 12886 | -0.135 | -0.0905 | No |
| 78 | Slc7a5 | 13021 | -0.143 | -0.0910 | No |
| 79 | Parn | 13120 | -0.148 | -0.0893 | No |
| 80 | Eif2s1 | 13234 | -0.153 | -0.0881 | No |
| 81 | Sec31a | 13513 | -0.167 | -0.0952 | No |
| 82 | Srprb | 13590 | -0.171 | -0.0912 | No |
| 83 | Fus | 13605 | -0.172 | -0.0837 | No |
| 84 | Sec11a | 13751 | -0.181 | -0.0829 | No |
| 85 | Npm1 | 13943 | -0.189 | -0.0843 | No |
| 86 | Banf1 | 14011 | -0.193 | -0.0786 | No |
| 87 | Nolc1 | 14404 | -0.216 | -0.0897 | No |
| 88 | Atf3 | 14655 | -0.231 | -0.0923 | No |
| 89 | Cks1b | 14821 | -0.241 | -0.0897 | No |
| 90 | Edc4 | 15028 | -0.254 | -0.0888 | No |
| 91 | Eef2 | 15138 | -0.261 | -0.0822 | No |
| 92 | Lsm4 | 15182 | -0.263 | -0.0719 | No |
| 93 | Slc1a4 | 15186 | -0.264 | -0.0594 | No |
| 94 | Eif2ak3 | 15600 | -0.288 | -0.0682 | No |
| 95 | Exosc4 | 15821 | -0.304 | -0.0656 | No |
| 96 | Nfyb | 16003 | -0.311 | -0.0606 | No |
| 97 | Exosc1 | 16008 | -0.312 | -0.0458 | No |
| 98 | Tatdn2 | 16455 | -0.345 | -0.0536 | No |
| 99 | Atp6v0d1 | 17019 | -0.400 | -0.0652 | No |
| 100 | Herpud1 | 17690 | -0.503 | -0.0776 | No |
| 101 | Rps14 | 17727 | -0.509 | -0.0552 | No |
| 102 | Lsm1 | 18094 | -0.605 | -0.0461 | No |
| 103 | Ero1a | 18143 | -0.628 | -0.0186 | No |
| 104 | Exosc5 | 18238 | -0.688 | 0.0093 | No |