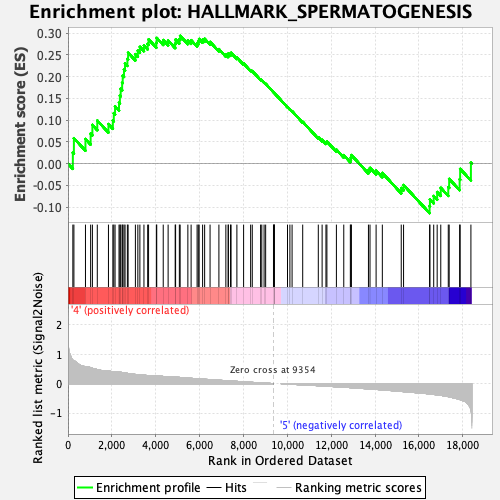
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.29394814 |
| Normalized Enrichment Score (NES) | 1.219115 |
| Nominal p-value | 0.16732283 |
| FDR q-value | 0.28442472 |
| FWER p-Value | 0.883 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gmcl1 | 221 | 0.796 | 0.0251 | Yes |
| 2 | Art3 | 269 | 0.771 | 0.0584 | Yes |
| 3 | Aurka | 796 | 0.578 | 0.0566 | Yes |
| 4 | Nefh | 1032 | 0.543 | 0.0691 | Yes |
| 5 | Nek2 | 1112 | 0.521 | 0.0891 | Yes |
| 6 | Cdkn3 | 1335 | 0.475 | 0.0991 | Yes |
| 7 | Rfc4 | 1844 | 0.421 | 0.0910 | Yes |
| 8 | Cnih2 | 2045 | 0.411 | 0.0992 | Yes |
| 9 | Oaz3 | 2088 | 0.409 | 0.1160 | Yes |
| 10 | Zc2hc1c | 2145 | 0.404 | 0.1318 | Yes |
| 11 | Ezh2 | 2326 | 0.393 | 0.1403 | Yes |
| 12 | Dcc | 2366 | 0.388 | 0.1562 | Yes |
| 13 | Dbf4 | 2402 | 0.384 | 0.1722 | Yes |
| 14 | Ttk | 2468 | 0.378 | 0.1863 | Yes |
| 15 | Acrbp | 2494 | 0.373 | 0.2023 | Yes |
| 16 | Strbp | 2552 | 0.368 | 0.2163 | Yes |
| 17 | Chfr | 2600 | 0.361 | 0.2306 | Yes |
| 18 | Zpbp | 2707 | 0.348 | 0.2410 | Yes |
| 19 | She | 2740 | 0.344 | 0.2553 | Yes |
| 20 | Tulp2 | 3072 | 0.312 | 0.2518 | Yes |
| 21 | Ide | 3184 | 0.302 | 0.2598 | Yes |
| 22 | Scg5 | 3277 | 0.298 | 0.2687 | Yes |
| 23 | Ccnb2 | 3462 | 0.285 | 0.2719 | Yes |
| 24 | Cct6b | 3632 | 0.273 | 0.2755 | Yes |
| 25 | Topbp1 | 3673 | 0.270 | 0.2859 | Yes |
| 26 | Stam2 | 4027 | 0.252 | 0.2783 | Yes |
| 27 | Pomc | 4045 | 0.251 | 0.2891 | Yes |
| 28 | Grm8 | 4341 | 0.243 | 0.2843 | Yes |
| 29 | Nf2 | 4564 | 0.238 | 0.2833 | Yes |
| 30 | Gfi1 | 4886 | 0.224 | 0.2762 | Yes |
| 31 | Mlf1 | 4905 | 0.222 | 0.2856 | Yes |
| 32 | Kif2c | 5076 | 0.211 | 0.2861 | Yes |
| 33 | Ncaph | 5112 | 0.208 | 0.2939 | Yes |
| 34 | Tnni3 | 5464 | 0.188 | 0.2836 | No |
| 35 | Hspa4l | 5612 | 0.179 | 0.2839 | No |
| 36 | Pias2 | 5885 | 0.165 | 0.2767 | No |
| 37 | Braf | 5938 | 0.163 | 0.2815 | No |
| 38 | Arl4a | 5978 | 0.160 | 0.2868 | No |
| 39 | Sirt1 | 6136 | 0.152 | 0.2853 | No |
| 40 | Agfg1 | 6229 | 0.147 | 0.2871 | No |
| 41 | Nphp1 | 6476 | 0.134 | 0.2800 | No |
| 42 | Ip6k1 | 6879 | 0.115 | 0.2634 | No |
| 43 | Rad17 | 7196 | 0.100 | 0.2508 | No |
| 44 | Zc3h14 | 7301 | 0.094 | 0.2495 | No |
| 45 | Tle4 | 7310 | 0.094 | 0.2534 | No |
| 46 | Camk4 | 7409 | 0.090 | 0.2523 | No |
| 47 | Mllt10 | 7433 | 0.089 | 0.2551 | No |
| 48 | Clpb | 7699 | 0.076 | 0.2442 | No |
| 49 | Adad1 | 8005 | 0.060 | 0.2304 | No |
| 50 | Csnk2a2 | 8326 | 0.046 | 0.2150 | No |
| 51 | Pacrg | 8400 | 0.042 | 0.2130 | No |
| 52 | Vdac3 | 8779 | 0.024 | 0.1935 | No |
| 53 | Lpin1 | 8834 | 0.023 | 0.1916 | No |
| 54 | Dmc1 | 8939 | 0.018 | 0.1868 | No |
| 55 | Coil | 9010 | 0.015 | 0.1837 | No |
| 56 | Hspa1l | 9367 | 0.000 | 0.1642 | No |
| 57 | Slc2a5 | 9396 | 0.000 | 0.1627 | No |
| 58 | Nos1 | 9398 | 0.000 | 0.1627 | No |
| 59 | Gsg1 | 9403 | 0.000 | 0.1624 | No |
| 60 | Gapdhs | 10006 | -0.012 | 0.1301 | No |
| 61 | Tsn | 10115 | -0.016 | 0.1250 | No |
| 62 | Prkar2a | 10215 | -0.021 | 0.1206 | No |
| 63 | Ybx2 | 10699 | -0.041 | 0.0961 | No |
| 64 | Phkg2 | 11410 | -0.069 | 0.0606 | No |
| 65 | Jam3 | 11584 | -0.076 | 0.0547 | No |
| 66 | Bub1 | 11759 | -0.084 | 0.0491 | No |
| 67 | Adam2 | 11802 | -0.086 | 0.0508 | No |
| 68 | Pgs1 | 12235 | -0.104 | 0.0321 | No |
| 69 | Mast2 | 12571 | -0.121 | 0.0194 | No |
| 70 | Pebp1 | 12871 | -0.134 | 0.0094 | No |
| 71 | Slc12a2 | 12896 | -0.135 | 0.0144 | No |
| 72 | Phf7 | 12918 | -0.137 | 0.0196 | No |
| 73 | Septin4 | 13696 | -0.177 | -0.0145 | No |
| 74 | Map7 | 13767 | -0.182 | -0.0099 | No |
| 75 | Spata6 | 14045 | -0.195 | -0.0159 | No |
| 76 | Mtor | 14329 | -0.211 | -0.0215 | No |
| 77 | Il12rb2 | 15191 | -0.264 | -0.0562 | No |
| 78 | Hspa2 | 15297 | -0.271 | -0.0493 | No |
| 79 | Ift88 | 16483 | -0.349 | -0.0977 | No |
| 80 | Ddx4 | 16499 | -0.350 | -0.0823 | No |
| 81 | Tcp11 | 16671 | -0.365 | -0.0746 | No |
| 82 | Dnajb8 | 16829 | -0.380 | -0.0654 | No |
| 83 | Taldo1 | 16993 | -0.397 | -0.0558 | No |
| 84 | Psmg1 | 17335 | -0.441 | -0.0539 | No |
| 85 | Cdk1 | 17376 | -0.446 | -0.0352 | No |
| 86 | Parp2 | 17853 | -0.536 | -0.0363 | No |
| 87 | Pcsk4 | 17877 | -0.542 | -0.0122 | No |
| 88 | Gstm5 | 18367 | -0.883 | 0.0022 | No |