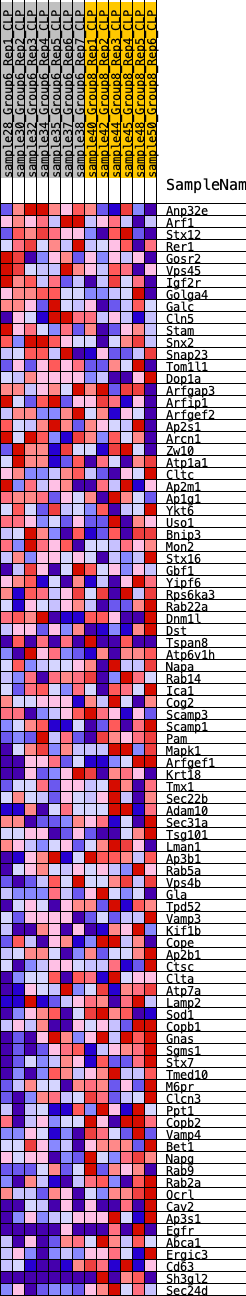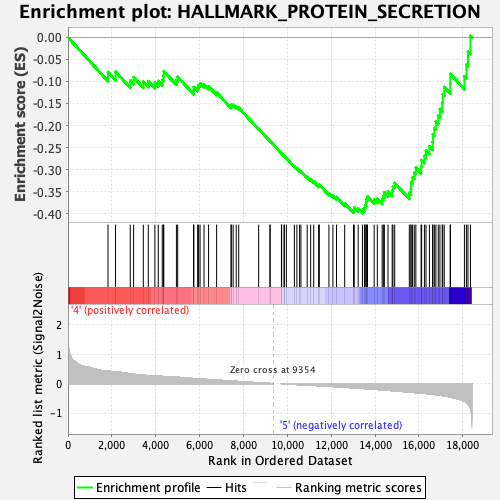
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.3989757 |
| Normalized Enrichment Score (NES) | -1.7543952 |
| Nominal p-value | 0.002105263 |
| FDR q-value | 0.06326781 |
| FWER p-Value | 0.062 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Anp32e | 1823 | 0.422 | -0.0787 | No |
| 2 | Arf1 | 2166 | 0.401 | -0.0776 | No |
| 3 | Stx12 | 2838 | 0.333 | -0.0978 | No |
| 4 | Rer1 | 2991 | 0.318 | -0.0904 | No |
| 5 | Gosr2 | 3433 | 0.288 | -0.1003 | No |
| 6 | Vps45 | 3660 | 0.271 | -0.0993 | No |
| 7 | Igf2r | 3961 | 0.256 | -0.1030 | No |
| 8 | Golga4 | 4114 | 0.251 | -0.0989 | No |
| 9 | Galc | 4297 | 0.247 | -0.0967 | No |
| 10 | Cln5 | 4353 | 0.242 | -0.0878 | No |
| 11 | Stam | 4367 | 0.241 | -0.0766 | No |
| 12 | Snx2 | 4942 | 0.219 | -0.0971 | No |
| 13 | Snap23 | 5001 | 0.216 | -0.0896 | No |
| 14 | Tom1l1 | 5721 | 0.174 | -0.1203 | No |
| 15 | Dop1a | 5737 | 0.173 | -0.1126 | No |
| 16 | Arfgap3 | 5910 | 0.164 | -0.1139 | No |
| 17 | Arfip1 | 5953 | 0.162 | -0.1082 | No |
| 18 | Arfgef2 | 6020 | 0.158 | -0.1040 | No |
| 19 | Ap2s1 | 6199 | 0.149 | -0.1064 | No |
| 20 | Arcn1 | 6408 | 0.139 | -0.1109 | No |
| 21 | Zw10 | 6775 | 0.120 | -0.1249 | No |
| 22 | Atp1a1 | 7432 | 0.089 | -0.1564 | No |
| 23 | Cltc | 7443 | 0.088 | -0.1526 | No |
| 24 | Ap2m1 | 7528 | 0.083 | -0.1531 | No |
| 25 | Ap1g1 | 7668 | 0.077 | -0.1569 | No |
| 26 | Ykt6 | 7782 | 0.072 | -0.1595 | No |
| 27 | Uso1 | 8691 | 0.029 | -0.2076 | No |
| 28 | Bnip3 | 9208 | 0.008 | -0.2354 | No |
| 29 | Mon2 | 9212 | 0.007 | -0.2352 | No |
| 30 | Stx16 | 9729 | -0.001 | -0.2634 | No |
| 31 | Gbf1 | 9739 | -0.001 | -0.2638 | No |
| 32 | Yipf6 | 9850 | -0.006 | -0.2695 | No |
| 33 | Rps6ka3 | 9860 | -0.006 | -0.2697 | No |
| 34 | Rab22a | 9954 | -0.010 | -0.2743 | No |
| 35 | Dnm1l | 10319 | -0.025 | -0.2929 | No |
| 36 | Dst | 10434 | -0.030 | -0.2976 | No |
| 37 | Tspan8 | 10557 | -0.035 | -0.3026 | No |
| 38 | Atp6v1h | 10619 | -0.038 | -0.3040 | No |
| 39 | Napa | 10904 | -0.049 | -0.3171 | No |
| 40 | Rab14 | 11062 | -0.056 | -0.3228 | No |
| 41 | Ica1 | 11201 | -0.061 | -0.3274 | No |
| 42 | Cog2 | 11424 | -0.070 | -0.3360 | No |
| 43 | Scamp3 | 11456 | -0.071 | -0.3342 | No |
| 44 | Scamp1 | 11895 | -0.089 | -0.3537 | No |
| 45 | Pam | 12077 | -0.098 | -0.3588 | No |
| 46 | Mapk1 | 12239 | -0.105 | -0.3624 | No |
| 47 | Arfgef1 | 12613 | -0.123 | -0.3767 | No |
| 48 | Krt18 | 13014 | -0.142 | -0.3915 | Yes |
| 49 | Tmx1 | 13049 | -0.144 | -0.3863 | Yes |
| 50 | Sec22b | 13228 | -0.153 | -0.3884 | Yes |
| 51 | Adam10 | 13422 | -0.163 | -0.3909 | Yes |
| 52 | Sec31a | 13513 | -0.167 | -0.3876 | Yes |
| 53 | Tsg101 | 13542 | -0.168 | -0.3808 | Yes |
| 54 | Lman1 | 13588 | -0.171 | -0.3748 | Yes |
| 55 | Ap3b1 | 13601 | -0.172 | -0.3670 | Yes |
| 56 | Rab5a | 13654 | -0.174 | -0.3612 | Yes |
| 57 | Vps4b | 13957 | -0.190 | -0.3683 | Yes |
| 58 | Gla | 14100 | -0.199 | -0.3663 | Yes |
| 59 | Tpd52 | 14321 | -0.211 | -0.3679 | Yes |
| 60 | Vamp3 | 14375 | -0.214 | -0.3602 | Yes |
| 61 | Kif1b | 14421 | -0.217 | -0.3520 | Yes |
| 62 | Cope | 14588 | -0.227 | -0.3498 | Yes |
| 63 | Ap2b1 | 14778 | -0.238 | -0.3484 | Yes |
| 64 | Ctsc | 14813 | -0.241 | -0.3384 | Yes |
| 65 | Clta | 14893 | -0.246 | -0.3306 | Yes |
| 66 | Atp7a | 15559 | -0.285 | -0.3528 | Yes |
| 67 | Lamp2 | 15634 | -0.290 | -0.3425 | Yes |
| 68 | Sod1 | 15639 | -0.291 | -0.3284 | Yes |
| 69 | Copb1 | 15708 | -0.296 | -0.3175 | Yes |
| 70 | Gnas | 15785 | -0.301 | -0.3068 | Yes |
| 71 | Sgms1 | 15857 | -0.306 | -0.2956 | Yes |
| 72 | Stx7 | 16096 | -0.318 | -0.2929 | Yes |
| 73 | Tmed10 | 16121 | -0.320 | -0.2784 | Yes |
| 74 | M6pr | 16247 | -0.332 | -0.2688 | Yes |
| 75 | Clcn3 | 16320 | -0.336 | -0.2562 | Yes |
| 76 | Ppt1 | 16474 | -0.348 | -0.2473 | Yes |
| 77 | Copb2 | 16621 | -0.360 | -0.2376 | Yes |
| 78 | Vamp4 | 16633 | -0.361 | -0.2204 | Yes |
| 79 | Bet1 | 16700 | -0.368 | -0.2058 | Yes |
| 80 | Napg | 16770 | -0.376 | -0.1910 | Yes |
| 81 | Rab9 | 16878 | -0.385 | -0.1779 | Yes |
| 82 | Rab2a | 16954 | -0.393 | -0.1626 | Yes |
| 83 | Ocrl | 17059 | -0.404 | -0.1483 | Yes |
| 84 | Cav2 | 17082 | -0.408 | -0.1294 | Yes |
| 85 | Ap3s1 | 17151 | -0.415 | -0.1127 | Yes |
| 86 | Egfr | 17425 | -0.454 | -0.1051 | Yes |
| 87 | Abca1 | 17431 | -0.455 | -0.0830 | Yes |
| 88 | Ergic3 | 18071 | -0.600 | -0.0882 | Yes |
| 89 | Cd63 | 18162 | -0.637 | -0.0617 | Yes |
| 90 | Sh3gl2 | 18233 | -0.686 | -0.0317 | Yes |
| 91 | Sec24d | 18345 | -0.834 | 0.0034 | Yes |