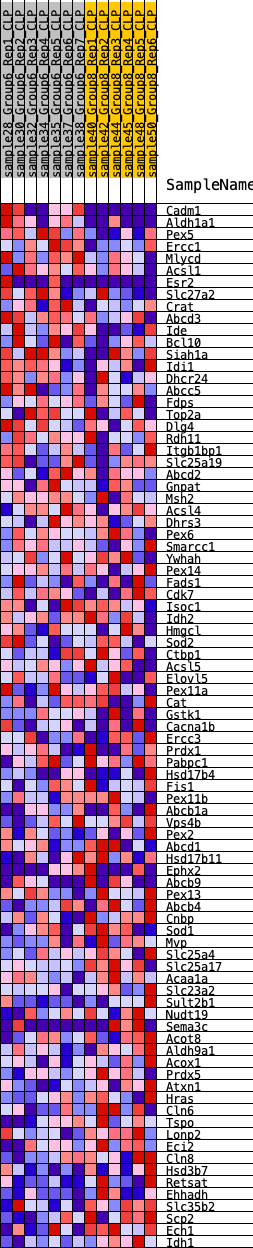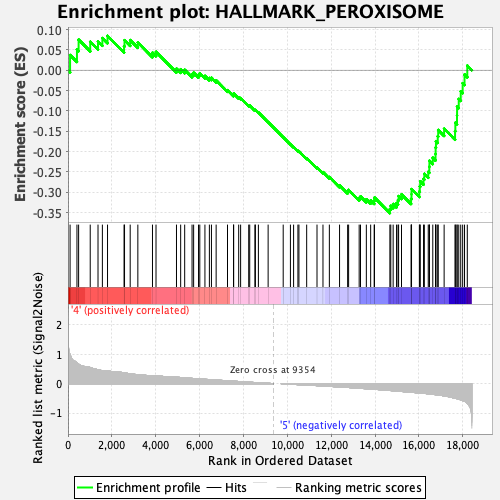
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.35115105 |
| Normalized Enrichment Score (NES) | -1.4777348 |
| Nominal p-value | 0.010660981 |
| FDR q-value | 0.21039888 |
| FWER p-Value | 0.4 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cadm1 | 95 | 0.984 | 0.0372 | No |
| 2 | Aldh1a1 | 408 | 0.708 | 0.0507 | No |
| 3 | Pex5 | 484 | 0.659 | 0.0750 | No |
| 4 | Ercc1 | 1013 | 0.546 | 0.0697 | No |
| 5 | Mlycd | 1368 | 0.471 | 0.0707 | No |
| 6 | Acsl1 | 1567 | 0.440 | 0.0789 | No |
| 7 | Esr2 | 1802 | 0.422 | 0.0843 | No |
| 8 | Slc27a2 | 2555 | 0.367 | 0.0591 | No |
| 9 | Crat | 2577 | 0.364 | 0.0736 | No |
| 10 | Abcd3 | 2835 | 0.333 | 0.0740 | No |
| 11 | Ide | 3184 | 0.302 | 0.0680 | No |
| 12 | Bcl10 | 3851 | 0.263 | 0.0430 | No |
| 13 | Siah1a | 4012 | 0.253 | 0.0452 | No |
| 14 | Idi1 | 4945 | 0.219 | 0.0037 | No |
| 15 | Dhcr24 | 5136 | 0.207 | 0.0023 | No |
| 16 | Abcc5 | 5319 | 0.197 | 0.0008 | No |
| 17 | Fdps | 5655 | 0.177 | -0.0098 | No |
| 18 | Top2a | 5727 | 0.174 | -0.0062 | No |
| 19 | Dlg4 | 5950 | 0.162 | -0.0113 | No |
| 20 | Rdh11 | 6008 | 0.159 | -0.0076 | No |
| 21 | Itgb1bp1 | 6242 | 0.146 | -0.0140 | No |
| 22 | Slc25a19 | 6445 | 0.136 | -0.0192 | No |
| 23 | Abcd2 | 6539 | 0.132 | -0.0186 | No |
| 24 | Gnpat | 6753 | 0.121 | -0.0250 | No |
| 25 | Msh2 | 7271 | 0.095 | -0.0491 | No |
| 26 | Acsl4 | 7545 | 0.083 | -0.0604 | No |
| 27 | Dhrs3 | 7552 | 0.082 | -0.0572 | No |
| 28 | Pex6 | 7781 | 0.072 | -0.0665 | No |
| 29 | Smarcc1 | 7869 | 0.068 | -0.0683 | No |
| 30 | Ywhah | 8238 | 0.049 | -0.0863 | No |
| 31 | Pex14 | 8284 | 0.047 | -0.0867 | No |
| 32 | Fads1 | 8520 | 0.036 | -0.0980 | No |
| 33 | Cdk7 | 8543 | 0.035 | -0.0977 | No |
| 34 | Isoc1 | 8679 | 0.029 | -0.1038 | No |
| 35 | Idh2 | 9123 | 0.011 | -0.1275 | No |
| 36 | Hmgcl | 9810 | -0.004 | -0.1648 | No |
| 37 | Sod2 | 10139 | -0.017 | -0.1820 | No |
| 38 | Ctbp1 | 10284 | -0.024 | -0.1888 | No |
| 39 | Acsl5 | 10481 | -0.032 | -0.1981 | No |
| 40 | Elovl5 | 10527 | -0.035 | -0.1991 | No |
| 41 | Pex11a | 10880 | -0.048 | -0.2162 | No |
| 42 | Cat | 11351 | -0.066 | -0.2390 | No |
| 43 | Gstk1 | 11623 | -0.078 | -0.2504 | No |
| 44 | Cacna1b | 11914 | -0.090 | -0.2624 | No |
| 45 | Ercc3 | 12379 | -0.111 | -0.2829 | No |
| 46 | Prdx1 | 12747 | -0.128 | -0.2974 | No |
| 47 | Pabpc1 | 12792 | -0.131 | -0.2942 | No |
| 48 | Hsd17b4 | 13273 | -0.155 | -0.3137 | No |
| 49 | Fis1 | 13334 | -0.159 | -0.3101 | No |
| 50 | Pex11b | 13600 | -0.172 | -0.3171 | No |
| 51 | Abcb1a | 13797 | -0.183 | -0.3199 | No |
| 52 | Vps4b | 13957 | -0.190 | -0.3204 | No |
| 53 | Pex2 | 13972 | -0.191 | -0.3129 | No |
| 54 | Abcd1 | 14673 | -0.231 | -0.3412 | Yes |
| 55 | Hsd17b11 | 14714 | -0.234 | -0.3333 | Yes |
| 56 | Ephx2 | 14822 | -0.241 | -0.3287 | Yes |
| 57 | Abcb9 | 14977 | -0.251 | -0.3263 | Yes |
| 58 | Pex13 | 15044 | -0.255 | -0.3189 | Yes |
| 59 | Abcb4 | 15071 | -0.257 | -0.3093 | Yes |
| 60 | Cnbp | 15203 | -0.265 | -0.3050 | Yes |
| 61 | Sod1 | 15639 | -0.291 | -0.3162 | Yes |
| 62 | Mvp | 15658 | -0.292 | -0.3046 | Yes |
| 63 | Slc25a4 | 15660 | -0.292 | -0.2921 | Yes |
| 64 | Slc25a17 | 16017 | -0.313 | -0.2981 | Yes |
| 65 | Acaa1a | 16036 | -0.314 | -0.2855 | Yes |
| 66 | Slc23a2 | 16056 | -0.315 | -0.2730 | Yes |
| 67 | Sult2b1 | 16207 | -0.329 | -0.2670 | Yes |
| 68 | Nudt19 | 16244 | -0.332 | -0.2546 | Yes |
| 69 | Sema3c | 16422 | -0.342 | -0.2496 | Yes |
| 70 | Acot8 | 16471 | -0.347 | -0.2372 | Yes |
| 71 | Aldh9a1 | 16476 | -0.348 | -0.2224 | Yes |
| 72 | Acox1 | 16629 | -0.360 | -0.2152 | Yes |
| 73 | Prdx5 | 16754 | -0.373 | -0.2058 | Yes |
| 74 | Atxn1 | 16759 | -0.374 | -0.1899 | Yes |
| 75 | Hras | 16774 | -0.376 | -0.1745 | Yes |
| 76 | Cln6 | 16855 | -0.382 | -0.1624 | Yes |
| 77 | Tspo | 16876 | -0.385 | -0.1469 | Yes |
| 78 | Lonp2 | 17146 | -0.414 | -0.1437 | Yes |
| 79 | Eci2 | 17645 | -0.492 | -0.1497 | Yes |
| 80 | Cln8 | 17658 | -0.495 | -0.1290 | Yes |
| 81 | Hsd3b7 | 17733 | -0.510 | -0.1111 | Yes |
| 82 | Retsat | 17737 | -0.510 | -0.0892 | Yes |
| 83 | Ehhadh | 17813 | -0.525 | -0.0707 | Yes |
| 84 | Slc35b2 | 17903 | -0.549 | -0.0519 | Yes |
| 85 | Scp2 | 17991 | -0.574 | -0.0319 | Yes |
| 86 | Ech1 | 18076 | -0.600 | -0.0106 | Yes |
| 87 | Idh1 | 18202 | -0.665 | 0.0112 | Yes |