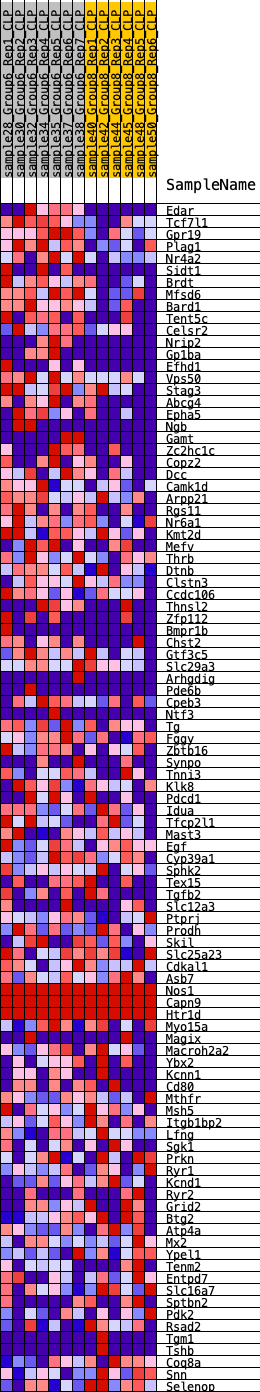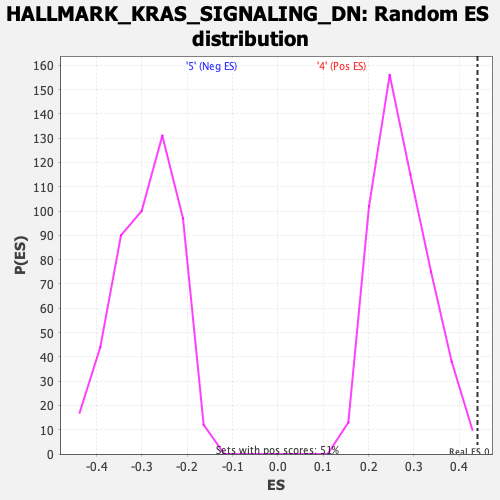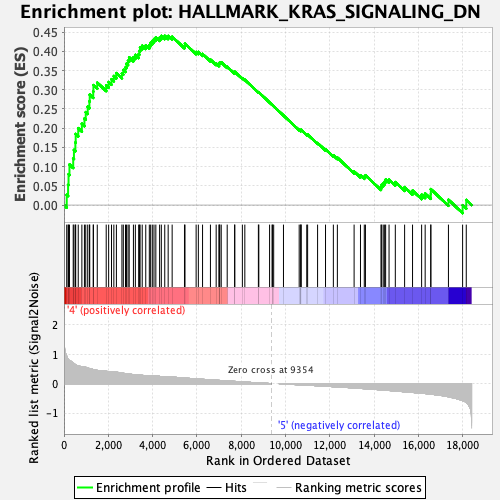
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.44027096 |
| Normalized Enrichment Score (NES) | 1.6086193 |
| Nominal p-value | 0.0019646366 |
| FDR q-value | 0.09419778 |
| FWER p-Value | 0.163 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Edar | 123 | 0.936 | 0.0265 | Yes |
| 2 | Tcf7l1 | 185 | 0.829 | 0.0526 | Yes |
| 3 | Gpr19 | 205 | 0.806 | 0.0801 | Yes |
| 4 | Plag1 | 247 | 0.779 | 0.1055 | Yes |
| 5 | Nr4a2 | 416 | 0.701 | 0.1212 | Yes |
| 6 | Sidt1 | 449 | 0.677 | 0.1435 | Yes |
| 7 | Brdt | 514 | 0.645 | 0.1629 | Yes |
| 8 | Mfsd6 | 528 | 0.641 | 0.1849 | Yes |
| 9 | Bard1 | 643 | 0.600 | 0.2000 | Yes |
| 10 | Tent5c | 807 | 0.575 | 0.2115 | Yes |
| 11 | Celsr2 | 923 | 0.565 | 0.2253 | Yes |
| 12 | Nrip2 | 982 | 0.554 | 0.2418 | Yes |
| 13 | Gp1ba | 1071 | 0.533 | 0.2559 | Yes |
| 14 | Efhd1 | 1147 | 0.513 | 0.2700 | Yes |
| 15 | Vps50 | 1162 | 0.509 | 0.2873 | Yes |
| 16 | Stag3 | 1320 | 0.477 | 0.2956 | Yes |
| 17 | Abcg4 | 1329 | 0.476 | 0.3121 | Yes |
| 18 | Epha5 | 1502 | 0.449 | 0.3186 | Yes |
| 19 | Ngb | 1906 | 0.418 | 0.3114 | Yes |
| 20 | Gamt | 2016 | 0.412 | 0.3201 | Yes |
| 21 | Zc2hc1c | 2145 | 0.404 | 0.3274 | Yes |
| 22 | Copz2 | 2251 | 0.398 | 0.3358 | Yes |
| 23 | Dcc | 2366 | 0.388 | 0.3433 | Yes |
| 24 | Camk1d | 2616 | 0.359 | 0.3425 | Yes |
| 25 | Arpp21 | 2678 | 0.352 | 0.3516 | Yes |
| 26 | Rgs11 | 2777 | 0.340 | 0.3583 | Yes |
| 27 | Nr6a1 | 2820 | 0.335 | 0.3679 | Yes |
| 28 | Kmt2d | 2892 | 0.327 | 0.3756 | Yes |
| 29 | Mefv | 2944 | 0.322 | 0.3843 | Yes |
| 30 | Thrb | 3135 | 0.307 | 0.3848 | Yes |
| 31 | Dtnb | 3222 | 0.300 | 0.3907 | Yes |
| 32 | Clstn3 | 3373 | 0.292 | 0.3929 | Yes |
| 33 | Ccdc106 | 3406 | 0.289 | 0.4014 | Yes |
| 34 | Thnsl2 | 3436 | 0.288 | 0.4100 | Yes |
| 35 | Zfp112 | 3532 | 0.281 | 0.4148 | Yes |
| 36 | Bmpr1b | 3692 | 0.270 | 0.4157 | Yes |
| 37 | Chst2 | 3853 | 0.263 | 0.4163 | Yes |
| 38 | Gtf3c5 | 3913 | 0.259 | 0.4223 | Yes |
| 39 | Slc29a3 | 3988 | 0.254 | 0.4272 | Yes |
| 40 | Arhgdig | 4064 | 0.251 | 0.4320 | Yes |
| 41 | Pde6b | 4147 | 0.249 | 0.4364 | Yes |
| 42 | Cpeb3 | 4318 | 0.245 | 0.4358 | Yes |
| 43 | Ntf3 | 4395 | 0.240 | 0.4402 | Yes |
| 44 | Tg | 4550 | 0.239 | 0.4403 | Yes |
| 45 | Fggy | 4704 | 0.229 | 0.4400 | No |
| 46 | Zbtb16 | 4884 | 0.224 | 0.4382 | No |
| 47 | Synpo | 5441 | 0.189 | 0.4145 | No |
| 48 | Tnni3 | 5464 | 0.188 | 0.4200 | No |
| 49 | Klk8 | 5967 | 0.161 | 0.3983 | No |
| 50 | Pdcd1 | 6067 | 0.155 | 0.3984 | No |
| 51 | Idua | 6257 | 0.145 | 0.3933 | No |
| 52 | Tfcp2l1 | 6613 | 0.128 | 0.3784 | No |
| 53 | Mast3 | 6872 | 0.116 | 0.3684 | No |
| 54 | Egf | 7002 | 0.109 | 0.3652 | No |
| 55 | Cyp39a1 | 7008 | 0.109 | 0.3688 | No |
| 56 | Sphk2 | 7035 | 0.108 | 0.3713 | No |
| 57 | Tex15 | 7100 | 0.105 | 0.3715 | No |
| 58 | Tgfb2 | 7370 | 0.092 | 0.3600 | No |
| 59 | Slc12a3 | 7707 | 0.075 | 0.3444 | No |
| 60 | Ptprj | 7711 | 0.075 | 0.3469 | No |
| 61 | Prodh | 8055 | 0.058 | 0.3302 | No |
| 62 | Skil | 8166 | 0.053 | 0.3260 | No |
| 63 | Slc25a23 | 8778 | 0.024 | 0.2935 | No |
| 64 | Cdkal1 | 8798 | 0.024 | 0.2933 | No |
| 65 | Asb7 | 9284 | 0.004 | 0.2670 | No |
| 66 | Nos1 | 9398 | 0.000 | 0.2608 | No |
| 67 | Capn9 | 9412 | 0.000 | 0.2601 | No |
| 68 | Htr1d | 9469 | 0.000 | 0.2570 | No |
| 69 | Myo15a | 9911 | -0.008 | 0.2332 | No |
| 70 | Magix | 10621 | -0.038 | 0.1959 | No |
| 71 | Macroh2a2 | 10695 | -0.041 | 0.1933 | No |
| 72 | Ybx2 | 10699 | -0.041 | 0.1946 | No |
| 73 | Kcnn1 | 10702 | -0.041 | 0.1960 | No |
| 74 | Cd80 | 10963 | -0.052 | 0.1836 | No |
| 75 | Mthfr | 11002 | -0.054 | 0.1834 | No |
| 76 | Msh5 | 11452 | -0.071 | 0.1614 | No |
| 77 | Itgb1bp2 | 11810 | -0.087 | 0.1450 | No |
| 78 | Lfng | 12164 | -0.102 | 0.1294 | No |
| 79 | Sgk1 | 12345 | -0.110 | 0.1234 | No |
| 80 | Prkn | 13101 | -0.147 | 0.0874 | No |
| 81 | Ryr1 | 13389 | -0.162 | 0.0775 | No |
| 82 | Kcnd1 | 13561 | -0.170 | 0.0742 | No |
| 83 | Ryr2 | 13613 | -0.173 | 0.0775 | No |
| 84 | Grid2 | 14315 | -0.210 | 0.0467 | No |
| 85 | Btg2 | 14346 | -0.212 | 0.0526 | No |
| 86 | Atp4a | 14416 | -0.216 | 0.0565 | No |
| 87 | Mx2 | 14480 | -0.221 | 0.0608 | No |
| 88 | Ypel1 | 14525 | -0.223 | 0.0664 | No |
| 89 | Tenm2 | 14679 | -0.232 | 0.0662 | No |
| 90 | Entpd7 | 14962 | -0.250 | 0.0597 | No |
| 91 | Slc16a7 | 15382 | -0.276 | 0.0466 | No |
| 92 | Sptbn2 | 15740 | -0.298 | 0.0377 | No |
| 93 | Pdk2 | 16150 | -0.324 | 0.0268 | No |
| 94 | Rsad2 | 16310 | -0.335 | 0.0300 | No |
| 95 | Tgm1 | 16564 | -0.356 | 0.0288 | No |
| 96 | Tshb | 16566 | -0.356 | 0.0414 | No |
| 97 | Coq8a | 17361 | -0.445 | 0.0138 | No |
| 98 | Snn | 18001 | -0.578 | -0.0006 | No |
| 99 | Selenop | 18166 | -0.641 | 0.0132 | No |