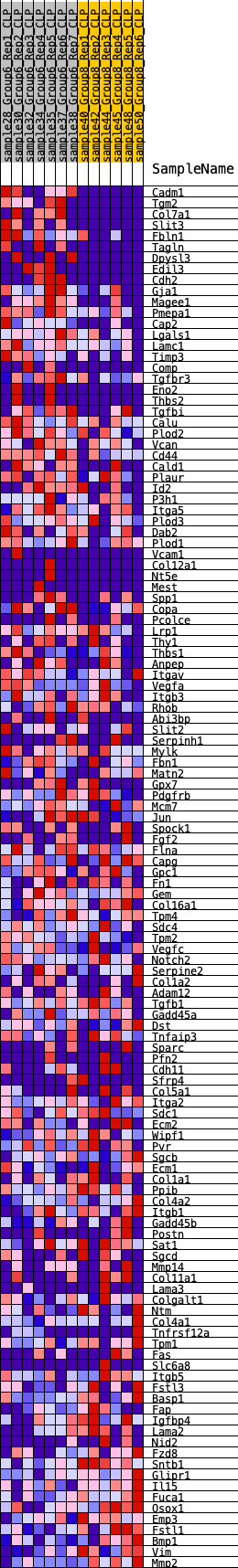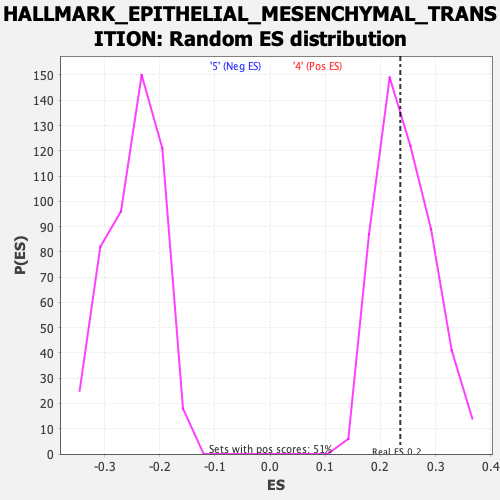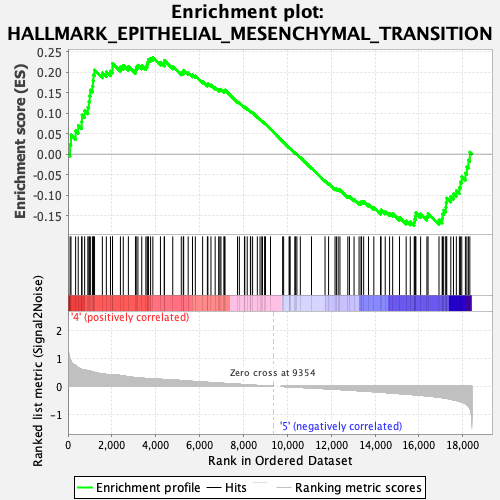
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 4 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.23604661 |
| Normalized Enrichment Score (NES) | 0.9657968 |
| Nominal p-value | 0.52362204 |
| FDR q-value | 0.7389809 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cadm1 | 95 | 0.984 | 0.0233 | Yes |
| 2 | Tgm2 | 136 | 0.895 | 0.0470 | Yes |
| 3 | Col7a1 | 347 | 0.746 | 0.0571 | Yes |
| 4 | Slit3 | 466 | 0.668 | 0.0700 | Yes |
| 5 | Fbln1 | 621 | 0.608 | 0.0792 | Yes |
| 6 | Tagln | 638 | 0.601 | 0.0957 | Yes |
| 7 | Dpysl3 | 759 | 0.582 | 0.1060 | Yes |
| 8 | Edil3 | 905 | 0.565 | 0.1144 | Yes |
| 9 | Cdh2 | 951 | 0.560 | 0.1282 | Yes |
| 10 | Gja1 | 987 | 0.554 | 0.1423 | Yes |
| 11 | Magee1 | 1017 | 0.546 | 0.1565 | Yes |
| 12 | Pmepa1 | 1113 | 0.520 | 0.1664 | Yes |
| 13 | Cap2 | 1142 | 0.514 | 0.1797 | Yes |
| 14 | Lgals1 | 1161 | 0.509 | 0.1935 | Yes |
| 15 | Lamc1 | 1210 | 0.500 | 0.2053 | Yes |
| 16 | Timp3 | 1566 | 0.440 | 0.1987 | Yes |
| 17 | Comp | 1747 | 0.427 | 0.2012 | Yes |
| 18 | Tgfbr3 | 1936 | 0.416 | 0.2029 | Yes |
| 19 | Eno2 | 2029 | 0.411 | 0.2098 | Yes |
| 20 | Thbs2 | 2030 | 0.411 | 0.2217 | Yes |
| 21 | Tgfbi | 2389 | 0.386 | 0.2133 | Yes |
| 22 | Calu | 2519 | 0.371 | 0.2170 | Yes |
| 23 | Plod2 | 2752 | 0.343 | 0.2142 | Yes |
| 24 | Vcan | 3075 | 0.311 | 0.2056 | Yes |
| 25 | Cd44 | 3109 | 0.309 | 0.2127 | Yes |
| 26 | Cald1 | 3187 | 0.302 | 0.2173 | Yes |
| 27 | Plaur | 3362 | 0.292 | 0.2162 | Yes |
| 28 | Id2 | 3550 | 0.279 | 0.2141 | Yes |
| 29 | P3h1 | 3608 | 0.275 | 0.2189 | Yes |
| 30 | Itga5 | 3640 | 0.273 | 0.2251 | Yes |
| 31 | Plod3 | 3666 | 0.271 | 0.2316 | Yes |
| 32 | Dab2 | 3764 | 0.269 | 0.2341 | Yes |
| 33 | Plod1 | 3868 | 0.262 | 0.2360 | Yes |
| 34 | Vcam1 | 4214 | 0.249 | 0.2244 | No |
| 35 | Col12a1 | 4391 | 0.240 | 0.2217 | No |
| 36 | Nt5e | 4392 | 0.240 | 0.2287 | No |
| 37 | Mest | 4775 | 0.227 | 0.2143 | No |
| 38 | Spp1 | 5164 | 0.205 | 0.1991 | No |
| 39 | Copa | 5254 | 0.200 | 0.2000 | No |
| 40 | Pcolce | 5273 | 0.199 | 0.2048 | No |
| 41 | Lrp1 | 5478 | 0.187 | 0.1991 | No |
| 42 | Thy1 | 5675 | 0.176 | 0.1934 | No |
| 43 | Thbs1 | 5804 | 0.169 | 0.1913 | No |
| 44 | Anpep | 6137 | 0.152 | 0.1776 | No |
| 45 | Itgav | 6353 | 0.142 | 0.1699 | No |
| 46 | Vegfa | 6377 | 0.140 | 0.1727 | No |
| 47 | Itgb3 | 6517 | 0.132 | 0.1689 | No |
| 48 | Rhob | 6708 | 0.123 | 0.1621 | No |
| 49 | Abi3bp | 6868 | 0.116 | 0.1568 | No |
| 50 | Slit2 | 6922 | 0.113 | 0.1571 | No |
| 51 | Serpinh1 | 6973 | 0.111 | 0.1576 | No |
| 52 | Mylk | 7107 | 0.105 | 0.1534 | No |
| 53 | Fbn1 | 7130 | 0.103 | 0.1551 | No |
| 54 | Matn2 | 7158 | 0.102 | 0.1566 | No |
| 55 | Gpx7 | 7727 | 0.074 | 0.1277 | No |
| 56 | Pdgfrb | 7811 | 0.071 | 0.1252 | No |
| 57 | Mcm7 | 8043 | 0.059 | 0.1143 | No |
| 58 | Jun | 8059 | 0.058 | 0.1151 | No |
| 59 | Spock1 | 8170 | 0.052 | 0.1106 | No |
| 60 | Fgf2 | 8315 | 0.046 | 0.1041 | No |
| 61 | Flna | 8327 | 0.046 | 0.1048 | No |
| 62 | Capg | 8415 | 0.041 | 0.1013 | No |
| 63 | Gpc1 | 8631 | 0.031 | 0.0904 | No |
| 64 | Fn1 | 8754 | 0.025 | 0.0844 | No |
| 65 | Gem | 8835 | 0.023 | 0.0807 | No |
| 66 | Col16a1 | 8867 | 0.021 | 0.0797 | No |
| 67 | Tpm4 | 8962 | 0.017 | 0.0750 | No |
| 68 | Sdc4 | 9003 | 0.016 | 0.0733 | No |
| 69 | Tpm2 | 9231 | 0.006 | 0.0610 | No |
| 70 | Vegfc | 9784 | -0.003 | 0.0309 | No |
| 71 | Notch2 | 9830 | -0.005 | 0.0286 | No |
| 72 | Serpine2 | 10078 | -0.015 | 0.0155 | No |
| 73 | Col1a2 | 10104 | -0.016 | 0.0146 | No |
| 74 | Adam12 | 10125 | -0.017 | 0.0140 | No |
| 75 | Tgfb1 | 10339 | -0.026 | 0.0031 | No |
| 76 | Gadd45a | 10377 | -0.028 | 0.0019 | No |
| 77 | Dst | 10434 | -0.030 | -0.0003 | No |
| 78 | Tnfaip3 | 10591 | -0.037 | -0.0078 | No |
| 79 | Sparc | 11100 | -0.058 | -0.0339 | No |
| 80 | Pfn2 | 11717 | -0.083 | -0.0652 | No |
| 81 | Cdh11 | 11879 | -0.089 | -0.0714 | No |
| 82 | Sfrp4 | 12173 | -0.102 | -0.0845 | No |
| 83 | Col5a1 | 12236 | -0.104 | -0.0848 | No |
| 84 | Itga2 | 12316 | -0.108 | -0.0860 | No |
| 85 | Sdc1 | 12396 | -0.112 | -0.0871 | No |
| 86 | Ecm2 | 12756 | -0.129 | -0.1030 | No |
| 87 | Wipf1 | 12830 | -0.132 | -0.1032 | No |
| 88 | Pvr | 13040 | -0.144 | -0.1104 | No |
| 89 | Sgcb | 13268 | -0.155 | -0.1184 | No |
| 90 | Ecm1 | 13356 | -0.160 | -0.1185 | No |
| 91 | Col1a1 | 13383 | -0.162 | -0.1152 | No |
| 92 | Ppib | 13478 | -0.166 | -0.1156 | No |
| 93 | Col4a2 | 13700 | -0.177 | -0.1225 | No |
| 94 | Itgb1 | 13942 | -0.189 | -0.1302 | No |
| 95 | Gadd45b | 14247 | -0.206 | -0.1409 | No |
| 96 | Postn | 14271 | -0.208 | -0.1361 | No |
| 97 | Sat1 | 14461 | -0.219 | -0.1401 | No |
| 98 | Sgcd | 14660 | -0.231 | -0.1443 | No |
| 99 | Mmp14 | 14800 | -0.240 | -0.1450 | No |
| 100 | Col11a1 | 15111 | -0.259 | -0.1544 | No |
| 101 | Lama3 | 15416 | -0.279 | -0.1629 | No |
| 102 | Colgalt1 | 15604 | -0.288 | -0.1648 | No |
| 103 | Ntm | 15783 | -0.301 | -0.1659 | No |
| 104 | Col4a1 | 15807 | -0.303 | -0.1584 | No |
| 105 | Tnfrsf12a | 15846 | -0.305 | -0.1516 | No |
| 106 | Tpm1 | 15856 | -0.306 | -0.1432 | No |
| 107 | Fas | 16074 | -0.317 | -0.1459 | No |
| 108 | Slc6a8 | 16361 | -0.340 | -0.1518 | No |
| 109 | Itgb5 | 16413 | -0.342 | -0.1446 | No |
| 110 | Fstl3 | 16912 | -0.390 | -0.1606 | No |
| 111 | Basp1 | 17056 | -0.404 | -0.1567 | No |
| 112 | Fap | 17076 | -0.407 | -0.1460 | No |
| 113 | Igfbp4 | 17125 | -0.412 | -0.1367 | No |
| 114 | Lama2 | 17223 | -0.425 | -0.1297 | No |
| 115 | Nid2 | 17235 | -0.426 | -0.1180 | No |
| 116 | Fzd8 | 17266 | -0.430 | -0.1072 | No |
| 117 | Sntb1 | 17451 | -0.458 | -0.1040 | No |
| 118 | Glipr1 | 17572 | -0.481 | -0.0966 | No |
| 119 | Il15 | 17703 | -0.504 | -0.0891 | No |
| 120 | Fuca1 | 17842 | -0.532 | -0.0813 | No |
| 121 | Qsox1 | 17888 | -0.546 | -0.0679 | No |
| 122 | Emp3 | 17951 | -0.560 | -0.0551 | No |
| 123 | Fstl1 | 18116 | -0.611 | -0.0464 | No |
| 124 | Bmp1 | 18182 | -0.650 | -0.0311 | No |
| 125 | Vim | 18252 | -0.704 | -0.0145 | No |
| 126 | Mmp2 | 18318 | -0.795 | 0.0049 | No |