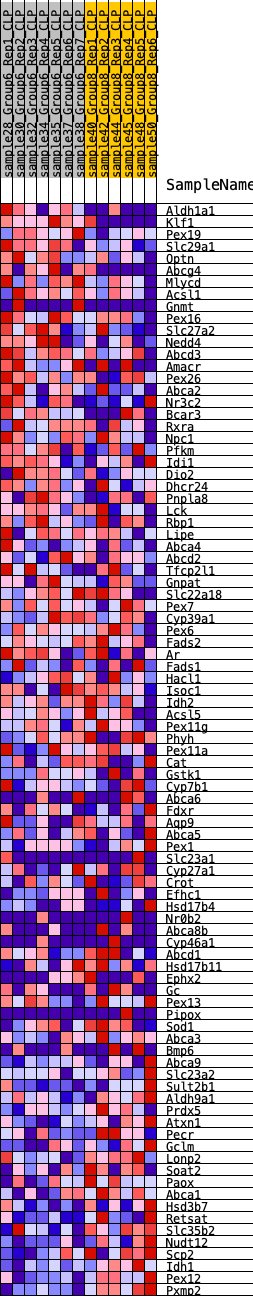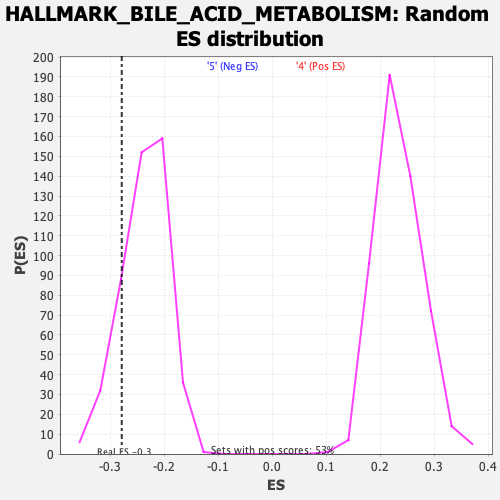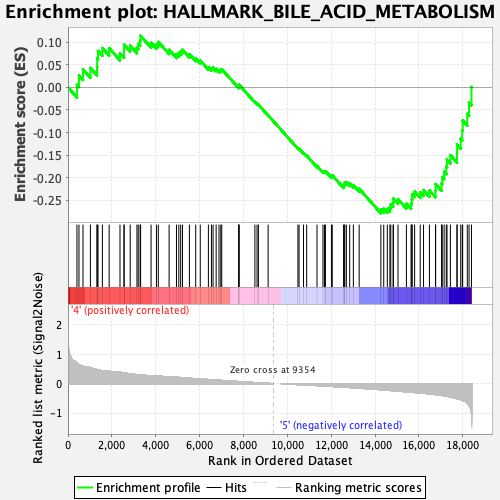
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group6_versus_Group8.Basophil_Pheno.cls #Group6_versus_Group8_repos |
| Phenotype | Basophil_Pheno.cls#Group6_versus_Group8_repos |
| Upregulated in class | 5 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.27884585 |
| Normalized Enrichment Score (NES) | -1.1798398 |
| Nominal p-value | 0.15611814 |
| FDR q-value | 0.43643156 |
| FWER p-Value | 0.933 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Aldh1a1 | 408 | 0.708 | 0.0057 | No |
| 2 | Klf1 | 500 | 0.652 | 0.0266 | No |
| 3 | Pex19 | 684 | 0.593 | 0.0400 | No |
| 4 | Slc29a1 | 1021 | 0.546 | 0.0433 | No |
| 5 | Optn | 1322 | 0.477 | 0.0458 | No |
| 6 | Abcg4 | 1329 | 0.476 | 0.0643 | No |
| 7 | Mlycd | 1368 | 0.471 | 0.0808 | No |
| 8 | Acsl1 | 1567 | 0.440 | 0.0874 | No |
| 9 | Gnmt | 1875 | 0.419 | 0.0873 | No |
| 10 | Pex16 | 2370 | 0.388 | 0.0756 | No |
| 11 | Slc27a2 | 2555 | 0.367 | 0.0801 | No |
| 12 | Nedd4 | 2556 | 0.367 | 0.0946 | No |
| 13 | Abcd3 | 2835 | 0.333 | 0.0926 | No |
| 14 | Amacr | 3143 | 0.306 | 0.0880 | No |
| 15 | Pex26 | 3201 | 0.301 | 0.0968 | No |
| 16 | Abca2 | 3286 | 0.297 | 0.1040 | No |
| 17 | Nr3c2 | 3310 | 0.295 | 0.1144 | No |
| 18 | Bcar3 | 3786 | 0.267 | 0.0990 | No |
| 19 | Rxra | 4032 | 0.251 | 0.0956 | No |
| 20 | Npc1 | 4117 | 0.250 | 0.1009 | No |
| 21 | Pfkm | 4609 | 0.235 | 0.0834 | No |
| 22 | Idi1 | 4945 | 0.219 | 0.0738 | No |
| 23 | Dio2 | 5052 | 0.213 | 0.0764 | No |
| 24 | Dhcr24 | 5136 | 0.207 | 0.0801 | No |
| 25 | Pnpla8 | 5220 | 0.202 | 0.0836 | No |
| 26 | Lck | 5534 | 0.184 | 0.0738 | No |
| 27 | Rbp1 | 5821 | 0.168 | 0.0648 | No |
| 28 | Lipe | 6029 | 0.157 | 0.0597 | No |
| 29 | Abca4 | 6400 | 0.139 | 0.0450 | No |
| 30 | Abcd2 | 6539 | 0.132 | 0.0427 | No |
| 31 | Tfcp2l1 | 6613 | 0.128 | 0.0438 | No |
| 32 | Gnpat | 6753 | 0.121 | 0.0410 | No |
| 33 | Slc22a18 | 6884 | 0.115 | 0.0384 | No |
| 34 | Pex7 | 6966 | 0.111 | 0.0384 | No |
| 35 | Cyp39a1 | 7008 | 0.109 | 0.0405 | No |
| 36 | Pex6 | 7781 | 0.072 | 0.0012 | No |
| 37 | Fads2 | 7794 | 0.071 | 0.0034 | No |
| 38 | Ar | 7796 | 0.071 | 0.0061 | No |
| 39 | Fads1 | 8520 | 0.036 | -0.0319 | No |
| 40 | Hacl1 | 8618 | 0.031 | -0.0359 | No |
| 41 | Isoc1 | 8679 | 0.029 | -0.0381 | No |
| 42 | Idh2 | 9123 | 0.011 | -0.0618 | No |
| 43 | Acsl5 | 10481 | -0.032 | -0.1346 | No |
| 44 | Pex11g | 10533 | -0.035 | -0.1360 | No |
| 45 | Phyh | 10736 | -0.043 | -0.1454 | No |
| 46 | Pex11a | 10880 | -0.048 | -0.1513 | No |
| 47 | Cat | 11351 | -0.066 | -0.1743 | No |
| 48 | Gstk1 | 11623 | -0.078 | -0.1860 | No |
| 49 | Cyp7b1 | 11691 | -0.081 | -0.1865 | No |
| 50 | Abca6 | 11735 | -0.083 | -0.1855 | No |
| 51 | Fdxr | 12009 | -0.095 | -0.1967 | No |
| 52 | Aqp9 | 12046 | -0.096 | -0.1948 | No |
| 53 | Abca5 | 12563 | -0.120 | -0.2182 | No |
| 54 | Pex1 | 12592 | -0.122 | -0.2149 | No |
| 55 | Slc23a1 | 12618 | -0.123 | -0.2114 | No |
| 56 | Cyp27a1 | 12689 | -0.127 | -0.2102 | No |
| 57 | Crot | 12841 | -0.133 | -0.2132 | No |
| 58 | Efhc1 | 13008 | -0.142 | -0.2166 | No |
| 59 | Hsd17b4 | 13273 | -0.155 | -0.2249 | No |
| 60 | Nr0b2 | 14262 | -0.207 | -0.2706 | Yes |
| 61 | Abca8b | 14393 | -0.215 | -0.2692 | Yes |
| 62 | Cyp46a1 | 14556 | -0.225 | -0.2691 | Yes |
| 63 | Abcd1 | 14673 | -0.231 | -0.2663 | Yes |
| 64 | Hsd17b11 | 14714 | -0.234 | -0.2592 | Yes |
| 65 | Ephx2 | 14822 | -0.241 | -0.2555 | Yes |
| 66 | Gc | 14832 | -0.242 | -0.2465 | Yes |
| 67 | Pex13 | 15044 | -0.255 | -0.2479 | Yes |
| 68 | Pipox | 15427 | -0.280 | -0.2577 | Yes |
| 69 | Sod1 | 15639 | -0.291 | -0.2577 | Yes |
| 70 | Abca3 | 15672 | -0.293 | -0.2478 | Yes |
| 71 | Bmp6 | 15690 | -0.294 | -0.2371 | Yes |
| 72 | Abca9 | 15801 | -0.302 | -0.2312 | Yes |
| 73 | Slc23a2 | 16056 | -0.315 | -0.2326 | Yes |
| 74 | Sult2b1 | 16207 | -0.329 | -0.2278 | Yes |
| 75 | Aldh9a1 | 16476 | -0.348 | -0.2286 | Yes |
| 76 | Prdx5 | 16754 | -0.373 | -0.2290 | Yes |
| 77 | Atxn1 | 16759 | -0.374 | -0.2144 | Yes |
| 78 | Pecr | 17029 | -0.401 | -0.2132 | Yes |
| 79 | Gclm | 17060 | -0.404 | -0.1988 | Yes |
| 80 | Lonp2 | 17146 | -0.414 | -0.1871 | Yes |
| 81 | Soat2 | 17243 | -0.427 | -0.1754 | Yes |
| 82 | Paox | 17277 | -0.431 | -0.1602 | Yes |
| 83 | Abca1 | 17431 | -0.455 | -0.1505 | Yes |
| 84 | Hsd3b7 | 17733 | -0.510 | -0.1467 | Yes |
| 85 | Retsat | 17737 | -0.510 | -0.1267 | Yes |
| 86 | Slc35b2 | 17903 | -0.549 | -0.1140 | Yes |
| 87 | Nudt12 | 17975 | -0.569 | -0.0954 | Yes |
| 88 | Scp2 | 17991 | -0.574 | -0.0735 | Yes |
| 89 | Idh1 | 18202 | -0.665 | -0.0586 | Yes |
| 90 | Pex12 | 18282 | -0.738 | -0.0337 | Yes |
| 91 | Pxmp2 | 18392 | -1.024 | 0.0009 | Yes |