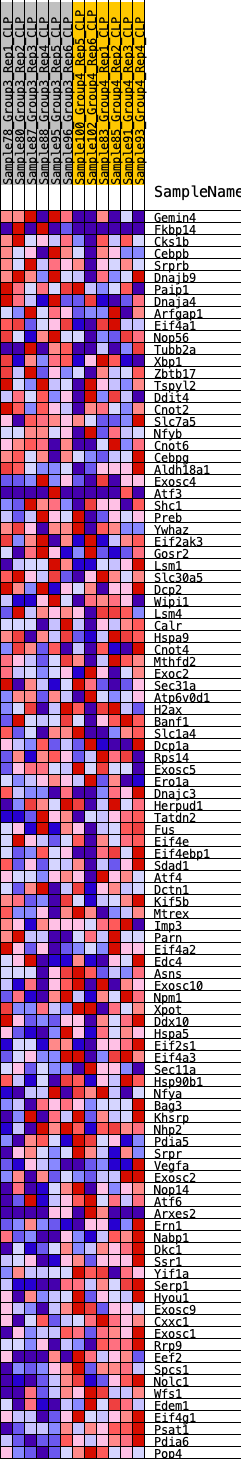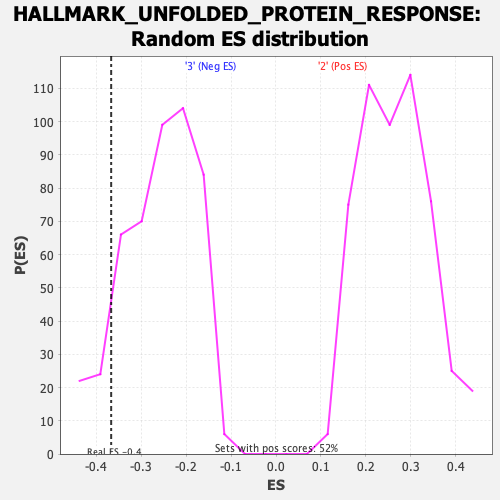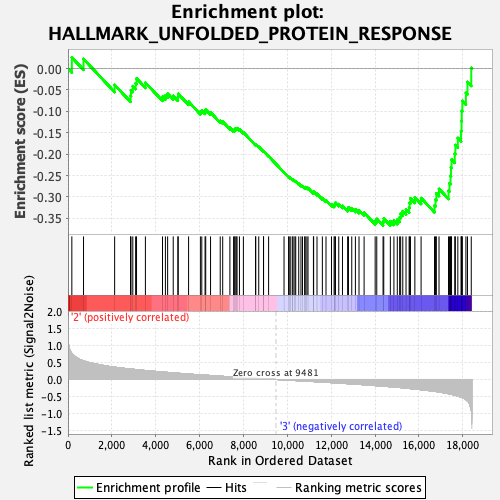
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | -0.36683124 |
| Normalized Enrichment Score (NES) | -1.4112703 |
| Nominal p-value | 0.101052634 |
| FDR q-value | 0.37768814 |
| FWER p-Value | 0.605 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gemin4 | 175 | 0.762 | 0.0259 | No |
| 2 | Fkbp14 | 708 | 0.548 | 0.0223 | No |
| 3 | Cks1b | 2126 | 0.362 | -0.0383 | No |
| 4 | Cebpb | 2850 | 0.306 | -0.0636 | No |
| 5 | Srprb | 2872 | 0.304 | -0.0506 | No |
| 6 | Dnajb9 | 2954 | 0.297 | -0.0412 | No |
| 7 | Paip1 | 3083 | 0.293 | -0.0346 | No |
| 8 | Dnaja4 | 3124 | 0.290 | -0.0233 | No |
| 9 | Arfgap1 | 3528 | 0.263 | -0.0331 | No |
| 10 | Eif4a1 | 4308 | 0.216 | -0.0656 | No |
| 11 | Nop56 | 4440 | 0.211 | -0.0630 | No |
| 12 | Tubb2a | 4542 | 0.206 | -0.0589 | No |
| 13 | Xbp1 | 4797 | 0.192 | -0.0638 | No |
| 14 | Zbtb17 | 5009 | 0.183 | -0.0669 | No |
| 15 | Tspyl2 | 5023 | 0.182 | -0.0591 | No |
| 16 | Ddit4 | 5497 | 0.158 | -0.0776 | No |
| 17 | Cnot2 | 6035 | 0.135 | -0.1007 | No |
| 18 | Slc7a5 | 6099 | 0.132 | -0.0980 | No |
| 19 | Nfyb | 6248 | 0.126 | -0.1002 | No |
| 20 | Cnot6 | 6278 | 0.125 | -0.0960 | No |
| 21 | Cebpg | 6498 | 0.115 | -0.1026 | No |
| 22 | Aldh18a1 | 6939 | 0.095 | -0.1222 | No |
| 23 | Exosc4 | 7052 | 0.091 | -0.1241 | No |
| 24 | Atf3 | 7380 | 0.081 | -0.1382 | No |
| 25 | Shc1 | 7548 | 0.074 | -0.1439 | No |
| 26 | Preb | 7601 | 0.073 | -0.1433 | No |
| 27 | Ywhaz | 7615 | 0.072 | -0.1407 | No |
| 28 | Eif2ak3 | 7669 | 0.070 | -0.1404 | No |
| 29 | Gosr2 | 7706 | 0.068 | -0.1392 | No |
| 30 | Lsm1 | 7815 | 0.064 | -0.1421 | No |
| 31 | Slc30a5 | 7995 | 0.058 | -0.1492 | No |
| 32 | Dcp2 | 8550 | 0.034 | -0.1778 | No |
| 33 | Wipi1 | 8558 | 0.034 | -0.1766 | No |
| 34 | Lsm4 | 8695 | 0.028 | -0.1828 | No |
| 35 | Calr | 8913 | 0.020 | -0.1937 | No |
| 36 | Hspa9 | 8914 | 0.020 | -0.1927 | No |
| 37 | Cnot4 | 9145 | 0.012 | -0.2048 | No |
| 38 | Mthfd2 | 9848 | -0.012 | -0.2426 | No |
| 39 | Exoc2 | 10054 | -0.019 | -0.2529 | No |
| 40 | Sec31a | 10085 | -0.019 | -0.2536 | No |
| 41 | Atp6v0d1 | 10156 | -0.022 | -0.2564 | No |
| 42 | H2ax | 10247 | -0.025 | -0.2602 | No |
| 43 | Banf1 | 10254 | -0.026 | -0.2593 | No |
| 44 | Slc1a4 | 10342 | -0.029 | -0.2627 | No |
| 45 | Dcp1a | 10381 | -0.031 | -0.2634 | No |
| 46 | Rps14 | 10518 | -0.036 | -0.2691 | No |
| 47 | Exosc5 | 10620 | -0.040 | -0.2728 | No |
| 48 | Ero1a | 10689 | -0.042 | -0.2745 | No |
| 49 | Dnajc3 | 10787 | -0.046 | -0.2777 | No |
| 50 | Herpud1 | 10814 | -0.047 | -0.2769 | No |
| 51 | Tatdn2 | 10889 | -0.050 | -0.2787 | No |
| 52 | Fus | 10941 | -0.051 | -0.2791 | No |
| 53 | Eif4e | 11188 | -0.059 | -0.2898 | No |
| 54 | Eif4ebp1 | 11198 | -0.060 | -0.2875 | No |
| 55 | Sdad1 | 11351 | -0.066 | -0.2927 | No |
| 56 | Atf4 | 11595 | -0.075 | -0.3025 | No |
| 57 | Dctn1 | 11761 | -0.082 | -0.3077 | No |
| 58 | Kif5b | 12011 | -0.091 | -0.3171 | No |
| 59 | Mtrex | 12130 | -0.096 | -0.3191 | No |
| 60 | Imp3 | 12166 | -0.097 | -0.3165 | No |
| 61 | Parn | 12192 | -0.098 | -0.3133 | No |
| 62 | Eif4a2 | 12343 | -0.104 | -0.3166 | No |
| 63 | Edc4 | 12512 | -0.112 | -0.3206 | No |
| 64 | Asns | 12747 | -0.122 | -0.3278 | No |
| 65 | Exosc10 | 12785 | -0.122 | -0.3241 | No |
| 66 | Npm1 | 12935 | -0.130 | -0.3262 | No |
| 67 | Xpot | 13101 | -0.137 | -0.3288 | No |
| 68 | Ddx10 | 13266 | -0.144 | -0.3311 | No |
| 69 | Hspa5 | 13498 | -0.154 | -0.3366 | No |
| 70 | Eif2s1 | 14002 | -0.178 | -0.3558 | No |
| 71 | Eif4a3 | 14075 | -0.182 | -0.3512 | No |
| 72 | Sec11a | 14362 | -0.197 | -0.3577 | Yes |
| 73 | Hsp90b1 | 14394 | -0.199 | -0.3501 | Yes |
| 74 | Nfya | 14694 | -0.212 | -0.3566 | Yes |
| 75 | Bag3 | 14857 | -0.222 | -0.3551 | Yes |
| 76 | Khsrp | 15011 | -0.231 | -0.3527 | Yes |
| 77 | Nhp2 | 15118 | -0.235 | -0.3476 | Yes |
| 78 | Pdia5 | 15164 | -0.238 | -0.3390 | Yes |
| 79 | Srpr | 15261 | -0.244 | -0.3329 | Yes |
| 80 | Vegfa | 15409 | -0.253 | -0.3292 | Yes |
| 81 | Exosc2 | 15550 | -0.263 | -0.3246 | Yes |
| 82 | Nop14 | 15573 | -0.264 | -0.3136 | Yes |
| 83 | Atf6 | 15611 | -0.267 | -0.3032 | Yes |
| 84 | Arxes2 | 15812 | -0.280 | -0.3011 | Yes |
| 85 | Ern1 | 16096 | -0.301 | -0.3025 | Yes |
| 86 | Nabp1 | 16708 | -0.347 | -0.3198 | Yes |
| 87 | Dkc1 | 16755 | -0.351 | -0.3060 | Yes |
| 88 | Ssr1 | 16792 | -0.354 | -0.2915 | Yes |
| 89 | Yif1a | 16913 | -0.364 | -0.2811 | Yes |
| 90 | Serp1 | 17346 | -0.418 | -0.2853 | Yes |
| 91 | Hyou1 | 17389 | -0.422 | -0.2680 | Yes |
| 92 | Exosc9 | 17437 | -0.429 | -0.2506 | Yes |
| 93 | Cxxc1 | 17458 | -0.434 | -0.2315 | Yes |
| 94 | Exosc1 | 17484 | -0.437 | -0.2126 | Yes |
| 95 | Rrp9 | 17631 | -0.462 | -0.1991 | Yes |
| 96 | Eef2 | 17658 | -0.469 | -0.1787 | Yes |
| 97 | Spcs1 | 17771 | -0.486 | -0.1622 | Yes |
| 98 | Nolc1 | 17917 | -0.519 | -0.1460 | Yes |
| 99 | Wfs1 | 17939 | -0.523 | -0.1228 | Yes |
| 100 | Edem1 | 17949 | -0.526 | -0.0989 | Yes |
| 101 | Eif4g1 | 17977 | -0.532 | -0.0756 | Yes |
| 102 | Psat1 | 18139 | -0.592 | -0.0569 | Yes |
| 103 | Pdia6 | 18210 | -0.630 | -0.0314 | Yes |
| 104 | Pop4 | 18383 | -0.908 | 0.0014 | Yes |