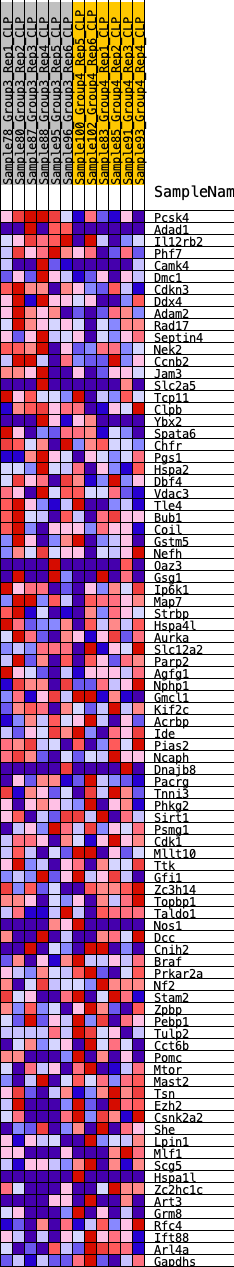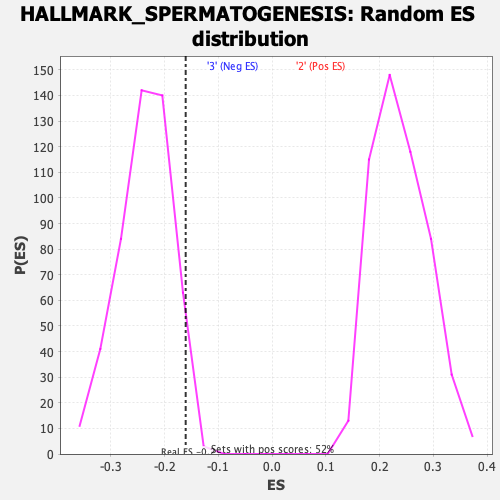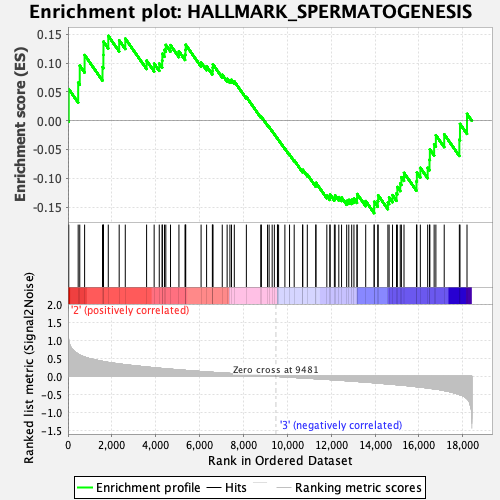
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.16066049 |
| Normalized Enrichment Score (NES) | -0.6775535 |
| Nominal p-value | 0.9752066 |
| FDR q-value | 0.9545248 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pcsk4 | 34 | 1.003 | 0.0549 | No |
| 2 | Adad1 | 464 | 0.614 | 0.0662 | No |
| 3 | Il12rb2 | 536 | 0.589 | 0.0957 | No |
| 4 | Phf7 | 757 | 0.539 | 0.1142 | No |
| 5 | Camk4 | 1573 | 0.416 | 0.0932 | No |
| 6 | Dmc1 | 1613 | 0.412 | 0.1144 | No |
| 7 | Cdkn3 | 1617 | 0.411 | 0.1375 | No |
| 8 | Ddx4 | 1836 | 0.386 | 0.1474 | No |
| 9 | Adam2 | 2333 | 0.341 | 0.1397 | No |
| 10 | Rad17 | 2611 | 0.322 | 0.1428 | No |
| 11 | Septin4 | 3583 | 0.258 | 0.1044 | No |
| 12 | Nek2 | 3927 | 0.240 | 0.0993 | No |
| 13 | Ccnb2 | 4160 | 0.226 | 0.0994 | No |
| 14 | Jam3 | 4292 | 0.217 | 0.1045 | No |
| 15 | Slc2a5 | 4300 | 0.217 | 0.1164 | No |
| 16 | Tcp11 | 4398 | 0.213 | 0.1232 | No |
| 17 | Clpb | 4459 | 0.209 | 0.1317 | No |
| 18 | Ybx2 | 4673 | 0.199 | 0.1314 | No |
| 19 | Spata6 | 5057 | 0.180 | 0.1207 | No |
| 20 | Chfr | 5336 | 0.166 | 0.1149 | No |
| 21 | Pgs1 | 5355 | 0.165 | 0.1233 | No |
| 22 | Hspa2 | 5367 | 0.164 | 0.1320 | No |
| 23 | Dbf4 | 6067 | 0.134 | 0.1014 | No |
| 24 | Vdac3 | 6318 | 0.123 | 0.0947 | No |
| 25 | Tle4 | 6581 | 0.112 | 0.0867 | No |
| 26 | Bub1 | 6597 | 0.111 | 0.0922 | No |
| 27 | Coil | 6605 | 0.111 | 0.0981 | No |
| 28 | Gstm5 | 7032 | 0.091 | 0.0800 | No |
| 29 | Nefh | 7254 | 0.086 | 0.0728 | No |
| 30 | Oaz3 | 7381 | 0.081 | 0.0705 | No |
| 31 | Gsg1 | 7454 | 0.078 | 0.0710 | No |
| 32 | Ip6k1 | 7581 | 0.074 | 0.0683 | No |
| 33 | Map7 | 8131 | 0.052 | 0.0413 | No |
| 34 | Strbp | 8787 | 0.025 | 0.0069 | No |
| 35 | Hspa4l | 8816 | 0.024 | 0.0067 | No |
| 36 | Aurka | 9096 | 0.013 | -0.0077 | No |
| 37 | Slc12a2 | 9168 | 0.011 | -0.0110 | No |
| 38 | Parp2 | 9307 | 0.006 | -0.0182 | No |
| 39 | Agfg1 | 9407 | 0.002 | -0.0235 | No |
| 40 | Nphp1 | 9548 | -0.001 | -0.0311 | No |
| 41 | Gmcl1 | 9554 | -0.001 | -0.0313 | No |
| 42 | Kif2c | 9600 | -0.002 | -0.0336 | No |
| 43 | Acrbp | 9887 | -0.013 | -0.0485 | No |
| 44 | Ide | 10099 | -0.020 | -0.0589 | No |
| 45 | Pias2 | 10311 | -0.028 | -0.0688 | No |
| 46 | Ncaph | 10693 | -0.042 | -0.0872 | No |
| 47 | Dnajb8 | 10703 | -0.043 | -0.0853 | No |
| 48 | Pacrg | 10909 | -0.050 | -0.0937 | No |
| 49 | Tnni3 | 11294 | -0.063 | -0.1110 | No |
| 50 | Phkg2 | 11304 | -0.064 | -0.1079 | No |
| 51 | Sirt1 | 11799 | -0.083 | -0.1302 | No |
| 52 | Psmg1 | 11938 | -0.088 | -0.1328 | No |
| 53 | Cdk1 | 11948 | -0.089 | -0.1282 | No |
| 54 | Mllt10 | 12143 | -0.096 | -0.1334 | No |
| 55 | Ttk | 12179 | -0.098 | -0.1298 | No |
| 56 | Gfi1 | 12345 | -0.104 | -0.1329 | No |
| 57 | Zc3h14 | 12470 | -0.110 | -0.1334 | No |
| 58 | Topbp1 | 12704 | -0.119 | -0.1394 | No |
| 59 | Taldo1 | 12800 | -0.123 | -0.1376 | No |
| 60 | Nos1 | 12932 | -0.130 | -0.1374 | No |
| 61 | Dcc | 13035 | -0.134 | -0.1354 | No |
| 62 | Cnih2 | 13174 | -0.140 | -0.1350 | No |
| 63 | Braf | 13182 | -0.140 | -0.1274 | No |
| 64 | Prkar2a | 13571 | -0.157 | -0.1397 | No |
| 65 | Nf2 | 13956 | -0.176 | -0.1507 | Yes |
| 66 | Stam2 | 13958 | -0.176 | -0.1408 | Yes |
| 67 | Zpbp | 14122 | -0.184 | -0.1393 | Yes |
| 68 | Pebp1 | 14138 | -0.185 | -0.1296 | Yes |
| 69 | Tulp2 | 14583 | -0.207 | -0.1421 | Yes |
| 70 | Cct6b | 14638 | -0.209 | -0.1332 | Yes |
| 71 | Pomc | 14794 | -0.218 | -0.1294 | Yes |
| 72 | Mtor | 14975 | -0.229 | -0.1262 | Yes |
| 73 | Mast2 | 15014 | -0.232 | -0.1152 | Yes |
| 74 | Tsn | 15146 | -0.236 | -0.1090 | Yes |
| 75 | Ezh2 | 15198 | -0.239 | -0.0982 | Yes |
| 76 | Csnk2a2 | 15317 | -0.247 | -0.0906 | Yes |
| 77 | She | 15886 | -0.286 | -0.1055 | Yes |
| 78 | Lpin1 | 15904 | -0.287 | -0.0901 | Yes |
| 79 | Mlf1 | 16062 | -0.298 | -0.0819 | Yes |
| 80 | Scg5 | 16396 | -0.321 | -0.0819 | Yes |
| 81 | Hspa1l | 16479 | -0.329 | -0.0677 | Yes |
| 82 | Zc2hc1c | 16496 | -0.330 | -0.0499 | Yes |
| 83 | Art3 | 16691 | -0.346 | -0.0409 | Yes |
| 84 | Grm8 | 16770 | -0.352 | -0.0252 | Yes |
| 85 | Rfc4 | 17150 | -0.393 | -0.0237 | Yes |
| 86 | Ift88 | 17840 | -0.502 | -0.0328 | Yes |
| 87 | Arl4a | 17869 | -0.509 | -0.0056 | Yes |
| 88 | Gapdhs | 18189 | -0.617 | 0.0120 | Yes |