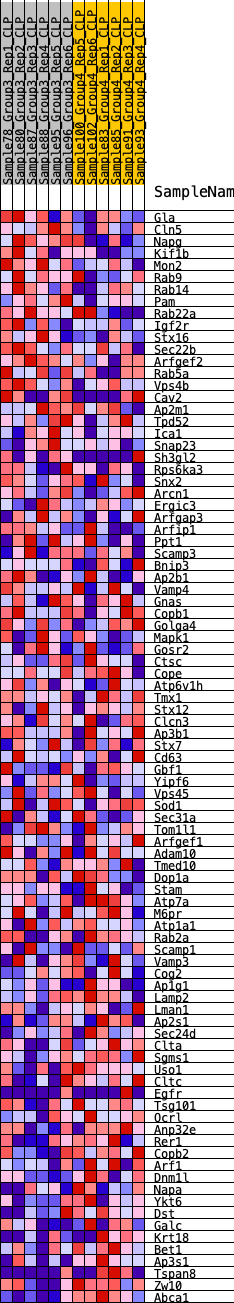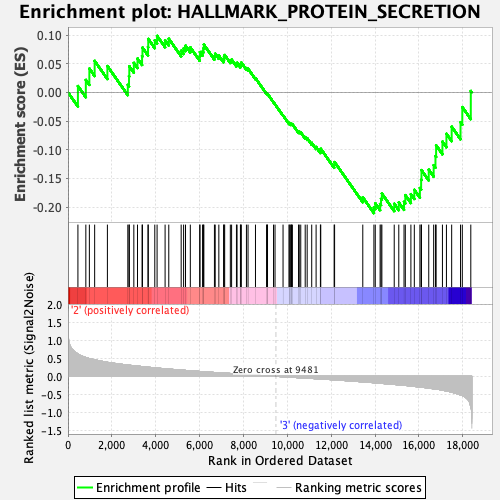
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.21043155 |
| Normalized Enrichment Score (NES) | -0.99512005 |
| Nominal p-value | 0.47316104 |
| FDR q-value | 0.9377753 |
| FWER p-Value | 0.993 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gla | 452 | 0.618 | 0.0111 | No |
| 2 | Cln5 | 813 | 0.525 | 0.0218 | No |
| 3 | Napg | 974 | 0.494 | 0.0416 | No |
| 4 | Kif1b | 1215 | 0.458 | 0.0549 | No |
| 5 | Mon2 | 1796 | 0.392 | 0.0459 | No |
| 6 | Rab9 | 2727 | 0.313 | 0.0133 | No |
| 7 | Rab14 | 2782 | 0.311 | 0.0283 | No |
| 8 | Pam | 2795 | 0.310 | 0.0456 | No |
| 9 | Rab22a | 3001 | 0.296 | 0.0515 | No |
| 10 | Igf2r | 3168 | 0.287 | 0.0590 | No |
| 11 | Stx16 | 3377 | 0.272 | 0.0634 | No |
| 12 | Sec22b | 3395 | 0.270 | 0.0781 | No |
| 13 | Arfgef2 | 3648 | 0.254 | 0.0790 | No |
| 14 | Rab5a | 3658 | 0.254 | 0.0932 | No |
| 15 | Vps4b | 3956 | 0.238 | 0.0908 | No |
| 16 | Cav2 | 4061 | 0.232 | 0.0985 | No |
| 17 | Ap2m1 | 4426 | 0.211 | 0.0909 | No |
| 18 | Tpd52 | 4594 | 0.203 | 0.0935 | No |
| 19 | Ica1 | 5159 | 0.175 | 0.0728 | No |
| 20 | Snap23 | 5270 | 0.169 | 0.0766 | No |
| 21 | Sh3gl2 | 5357 | 0.165 | 0.0815 | No |
| 22 | Rps6ka3 | 5576 | 0.154 | 0.0785 | No |
| 23 | Snx2 | 6006 | 0.136 | 0.0629 | No |
| 24 | Arcn1 | 6014 | 0.136 | 0.0704 | No |
| 25 | Ergic3 | 6134 | 0.131 | 0.0715 | No |
| 26 | Arfgap3 | 6174 | 0.130 | 0.0768 | No |
| 27 | Arfip1 | 6193 | 0.129 | 0.0833 | No |
| 28 | Ppt1 | 6675 | 0.108 | 0.0633 | No |
| 29 | Scamp3 | 6710 | 0.106 | 0.0676 | No |
| 30 | Bnip3 | 6876 | 0.098 | 0.0642 | No |
| 31 | Ap2b1 | 7093 | 0.089 | 0.0576 | No |
| 32 | Vamp4 | 7113 | 0.088 | 0.0616 | No |
| 33 | Gnas | 7142 | 0.087 | 0.0652 | No |
| 34 | Copb1 | 7400 | 0.081 | 0.0558 | No |
| 35 | Golga4 | 7458 | 0.078 | 0.0572 | No |
| 36 | Mapk1 | 7679 | 0.069 | 0.0492 | No |
| 37 | Gosr2 | 7706 | 0.068 | 0.0517 | No |
| 38 | Ctsc | 7860 | 0.063 | 0.0470 | No |
| 39 | Cope | 7876 | 0.062 | 0.0498 | No |
| 40 | Atp6v1h | 7899 | 0.061 | 0.0521 | No |
| 41 | Tmx1 | 8134 | 0.052 | 0.0424 | No |
| 42 | Stx12 | 8207 | 0.049 | 0.0413 | No |
| 43 | Clcn3 | 8546 | 0.034 | 0.0249 | No |
| 44 | Ap3b1 | 9052 | 0.015 | -0.0018 | No |
| 45 | Stx7 | 9076 | 0.014 | -0.0023 | No |
| 46 | Cd63 | 9095 | 0.013 | -0.0025 | No |
| 47 | Gbf1 | 9369 | 0.004 | -0.0172 | No |
| 48 | Yipf6 | 9437 | 0.001 | -0.0208 | No |
| 49 | Vps45 | 9803 | -0.010 | -0.0401 | No |
| 50 | Sod1 | 10080 | -0.019 | -0.0541 | No |
| 51 | Sec31a | 10085 | -0.019 | -0.0532 | No |
| 52 | Tom1l1 | 10151 | -0.022 | -0.0554 | No |
| 53 | Arfgef1 | 10175 | -0.023 | -0.0554 | No |
| 54 | Adam10 | 10188 | -0.023 | -0.0547 | No |
| 55 | Tmed10 | 10229 | -0.024 | -0.0555 | No |
| 56 | Dop1a | 10510 | -0.036 | -0.0687 | No |
| 57 | Stam | 10540 | -0.037 | -0.0681 | No |
| 58 | Atp7a | 10610 | -0.039 | -0.0696 | No |
| 59 | M6pr | 10813 | -0.047 | -0.0779 | No |
| 60 | Atp1a1 | 10903 | -0.050 | -0.0799 | No |
| 61 | Rab2a | 11110 | -0.056 | -0.0879 | No |
| 62 | Scamp1 | 11310 | -0.064 | -0.0951 | No |
| 63 | Vamp3 | 11506 | -0.071 | -0.1016 | No |
| 64 | Cog2 | 11522 | -0.072 | -0.0982 | No |
| 65 | Ap1g1 | 12132 | -0.096 | -0.1259 | No |
| 66 | Lamp2 | 12153 | -0.097 | -0.1214 | No |
| 67 | Lman1 | 13438 | -0.151 | -0.1828 | No |
| 68 | Ap2s1 | 13945 | -0.175 | -0.2003 | Yes |
| 69 | Sec24d | 14007 | -0.178 | -0.1933 | Yes |
| 70 | Clta | 14228 | -0.190 | -0.1943 | Yes |
| 71 | Sgms1 | 14270 | -0.192 | -0.1855 | Yes |
| 72 | Uso1 | 14308 | -0.194 | -0.1762 | Yes |
| 73 | Cltc | 14871 | -0.223 | -0.1941 | Yes |
| 74 | Egfr | 15079 | -0.235 | -0.1918 | Yes |
| 75 | Tsg101 | 15318 | -0.247 | -0.1905 | Yes |
| 76 | Ocrl | 15377 | -0.252 | -0.1791 | Yes |
| 77 | Anp32e | 15631 | -0.269 | -0.1774 | Yes |
| 78 | Rer1 | 15787 | -0.279 | -0.1697 | Yes |
| 79 | Copb2 | 16042 | -0.295 | -0.1665 | Yes |
| 80 | Arf1 | 16098 | -0.301 | -0.1521 | Yes |
| 81 | Dnm1l | 16114 | -0.302 | -0.1355 | Yes |
| 82 | Napa | 16445 | -0.326 | -0.1346 | Yes |
| 83 | Ykt6 | 16669 | -0.344 | -0.1269 | Yes |
| 84 | Dst | 16751 | -0.351 | -0.1110 | Yes |
| 85 | Galc | 16781 | -0.353 | -0.0922 | Yes |
| 86 | Krt18 | 17069 | -0.383 | -0.0857 | Yes |
| 87 | Bet1 | 17251 | -0.406 | -0.0721 | Yes |
| 88 | Ap3s1 | 17490 | -0.438 | -0.0598 | Yes |
| 89 | Tspan8 | 17894 | -0.515 | -0.0520 | Yes |
| 90 | Zw10 | 17975 | -0.532 | -0.0256 | Yes |
| 91 | Abca1 | 18362 | -0.851 | 0.0025 | Yes |