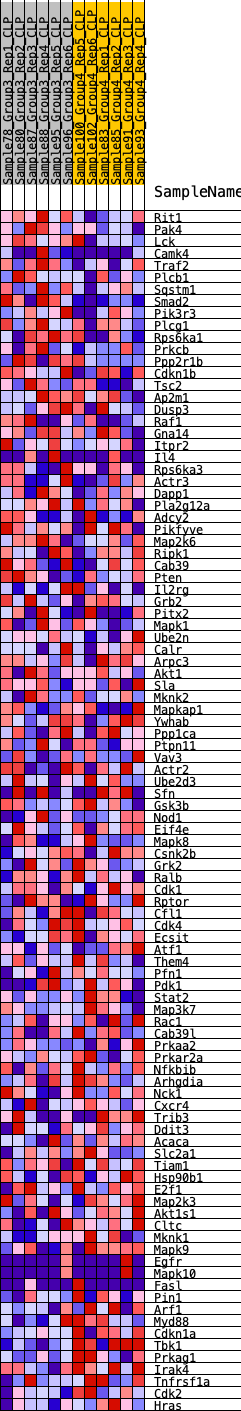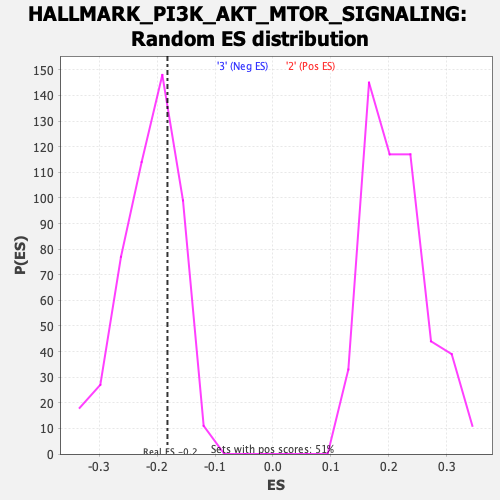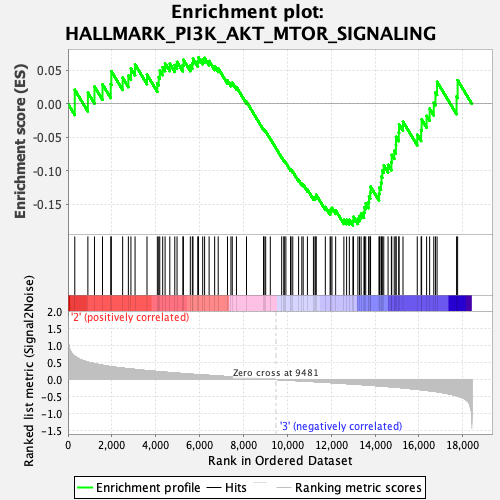
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | -0.18221934 |
| Normalized Enrichment Score (NES) | -0.8574174 |
| Nominal p-value | 0.67813766 |
| FDR q-value | 0.9307557 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rit1 | 309 | 0.680 | 0.0209 | No |
| 2 | Pak4 | 906 | 0.509 | 0.0167 | No |
| 3 | Lck | 1207 | 0.460 | 0.0258 | No |
| 4 | Camk4 | 1573 | 0.416 | 0.0290 | No |
| 5 | Traf2 | 1949 | 0.376 | 0.0294 | No |
| 6 | Plcb1 | 1975 | 0.373 | 0.0488 | No |
| 7 | Sqstm1 | 2491 | 0.332 | 0.0391 | No |
| 8 | Smad2 | 2752 | 0.312 | 0.0422 | No |
| 9 | Pik3r3 | 2869 | 0.305 | 0.0528 | No |
| 10 | Plcg1 | 3057 | 0.294 | 0.0589 | No |
| 11 | Rps6ka1 | 3601 | 0.257 | 0.0436 | No |
| 12 | Prkcb | 4073 | 0.231 | 0.0307 | No |
| 13 | Ppp2r1b | 4135 | 0.227 | 0.0400 | No |
| 14 | Cdkn1b | 4183 | 0.225 | 0.0499 | No |
| 15 | Tsc2 | 4316 | 0.216 | 0.0547 | No |
| 16 | Ap2m1 | 4426 | 0.211 | 0.0605 | No |
| 17 | Dusp3 | 4641 | 0.201 | 0.0599 | No |
| 18 | Raf1 | 4868 | 0.188 | 0.0581 | No |
| 19 | Gna14 | 4971 | 0.184 | 0.0627 | No |
| 20 | Itpr2 | 5236 | 0.171 | 0.0578 | No |
| 21 | Il4 | 5261 | 0.170 | 0.0659 | No |
| 22 | Rps6ka3 | 5576 | 0.154 | 0.0574 | No |
| 23 | Actr3 | 5668 | 0.150 | 0.0607 | No |
| 24 | Dapp1 | 5697 | 0.149 | 0.0675 | No |
| 25 | Pla2g12a | 5915 | 0.140 | 0.0634 | No |
| 26 | Adcy2 | 5950 | 0.139 | 0.0693 | No |
| 27 | Pikfyve | 6140 | 0.131 | 0.0663 | No |
| 28 | Map2k6 | 6227 | 0.127 | 0.0686 | No |
| 29 | Ripk1 | 6433 | 0.118 | 0.0640 | No |
| 30 | Cab39 | 6685 | 0.107 | 0.0563 | No |
| 31 | Pten | 6847 | 0.099 | 0.0530 | No |
| 32 | Il2rg | 7270 | 0.086 | 0.0347 | No |
| 33 | Grb2 | 7428 | 0.079 | 0.0305 | No |
| 34 | Pitx2 | 7488 | 0.077 | 0.0316 | No |
| 35 | Mapk1 | 7679 | 0.069 | 0.0251 | No |
| 36 | Ube2n | 8137 | 0.052 | 0.0030 | No |
| 37 | Calr | 8913 | 0.020 | -0.0382 | No |
| 38 | Arpc3 | 8940 | 0.019 | -0.0385 | No |
| 39 | Akt1 | 9011 | 0.016 | -0.0414 | No |
| 40 | Sla | 9222 | 0.009 | -0.0524 | No |
| 41 | Mknk2 | 9741 | -0.008 | -0.0802 | No |
| 42 | Mapkap1 | 9839 | -0.011 | -0.0849 | No |
| 43 | Ywhab | 9859 | -0.012 | -0.0853 | No |
| 44 | Ppp1ca | 9932 | -0.014 | -0.0884 | No |
| 45 | Ptpn11 | 10145 | -0.022 | -0.0988 | No |
| 46 | Vav3 | 10167 | -0.022 | -0.0987 | No |
| 47 | Actr2 | 10246 | -0.025 | -0.1016 | No |
| 48 | Ube2d3 | 10512 | -0.036 | -0.1140 | No |
| 49 | Sfn | 10659 | -0.041 | -0.1197 | No |
| 50 | Gsk3b | 10721 | -0.043 | -0.1207 | No |
| 51 | Nod1 | 10914 | -0.050 | -0.1283 | No |
| 52 | Eif4e | 11188 | -0.059 | -0.1400 | No |
| 53 | Mapk8 | 11258 | -0.062 | -0.1403 | No |
| 54 | Csnk2b | 11298 | -0.064 | -0.1389 | No |
| 55 | Grk2 | 11311 | -0.064 | -0.1360 | No |
| 56 | Ralb | 11730 | -0.080 | -0.1543 | No |
| 57 | Cdk1 | 11948 | -0.089 | -0.1613 | No |
| 58 | Rptor | 11980 | -0.090 | -0.1580 | No |
| 59 | Cfl1 | 12033 | -0.092 | -0.1557 | No |
| 60 | Cdk4 | 12201 | -0.098 | -0.1593 | No |
| 61 | Ecsit | 12576 | -0.114 | -0.1734 | No |
| 62 | Atf1 | 12700 | -0.119 | -0.1735 | No |
| 63 | Them4 | 12825 | -0.124 | -0.1734 | No |
| 64 | Pfn1 | 12987 | -0.131 | -0.1749 | Yes |
| 65 | Pdk1 | 13016 | -0.133 | -0.1691 | Yes |
| 66 | Stat2 | 13215 | -0.142 | -0.1720 | Yes |
| 67 | Map3k7 | 13286 | -0.145 | -0.1678 | Yes |
| 68 | Rac1 | 13362 | -0.148 | -0.1637 | Yes |
| 69 | Cab39l | 13492 | -0.154 | -0.1621 | Yes |
| 70 | Prkaa2 | 13517 | -0.155 | -0.1548 | Yes |
| 71 | Prkar2a | 13571 | -0.157 | -0.1490 | Yes |
| 72 | Nfkbib | 13705 | -0.165 | -0.1471 | Yes |
| 73 | Arhgdia | 13722 | -0.165 | -0.1388 | Yes |
| 74 | Nck1 | 13781 | -0.168 | -0.1326 | Yes |
| 75 | Cxcr4 | 13792 | -0.168 | -0.1238 | Yes |
| 76 | Trib3 | 14177 | -0.187 | -0.1344 | Yes |
| 77 | Ddit3 | 14199 | -0.189 | -0.1251 | Yes |
| 78 | Acaca | 14268 | -0.192 | -0.1181 | Yes |
| 79 | Slc2a1 | 14292 | -0.194 | -0.1086 | Yes |
| 80 | Tiam1 | 14329 | -0.196 | -0.0997 | Yes |
| 81 | Hsp90b1 | 14394 | -0.199 | -0.0922 | Yes |
| 82 | E2f1 | 14591 | -0.207 | -0.0914 | Yes |
| 83 | Map2k3 | 14741 | -0.215 | -0.0875 | Yes |
| 84 | Akt1s1 | 14758 | -0.216 | -0.0764 | Yes |
| 85 | Cltc | 14871 | -0.223 | -0.0702 | Yes |
| 86 | Mknk1 | 14949 | -0.227 | -0.0617 | Yes |
| 87 | Mapk9 | 14955 | -0.228 | -0.0493 | Yes |
| 88 | Egfr | 15079 | -0.235 | -0.0430 | Yes |
| 89 | Mapk10 | 15091 | -0.235 | -0.0306 | Yes |
| 90 | Fasl | 15271 | -0.245 | -0.0268 | Yes |
| 91 | Pin1 | 15921 | -0.288 | -0.0462 | Yes |
| 92 | Arf1 | 16098 | -0.301 | -0.0391 | Yes |
| 93 | Myd88 | 16120 | -0.303 | -0.0234 | Yes |
| 94 | Cdkn1a | 16347 | -0.318 | -0.0181 | Yes |
| 95 | Tbk1 | 16486 | -0.329 | -0.0073 | Yes |
| 96 | Prkag1 | 16674 | -0.345 | 0.0016 | Yes |
| 97 | Irak4 | 16750 | -0.351 | 0.0171 | Yes |
| 98 | Tnfrsf1a | 16824 | -0.357 | 0.0329 | Yes |
| 99 | Cdk2 | 17714 | -0.478 | 0.0110 | Yes |
| 100 | Hras | 17761 | -0.484 | 0.0353 | Yes |