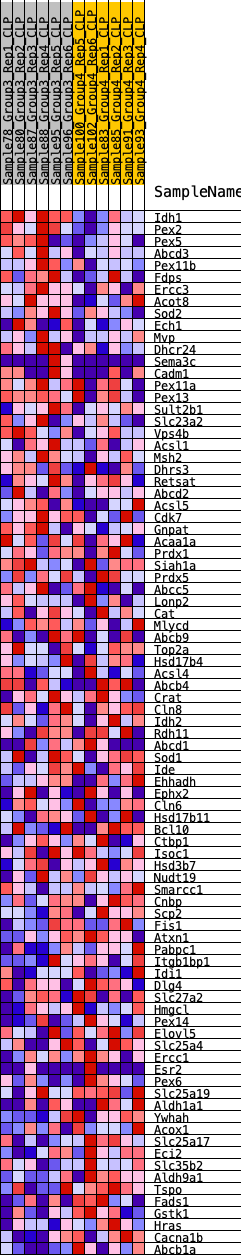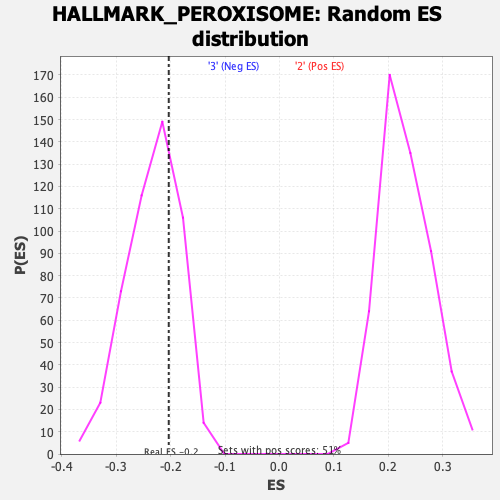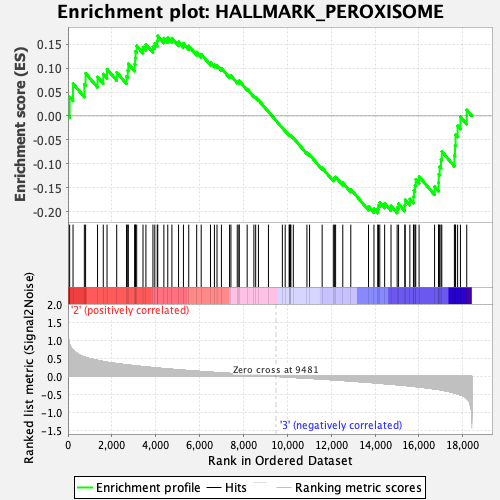
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.20344707 |
| Normalized Enrichment Score (NES) | -0.87461865 |
| Nominal p-value | 0.6899384 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idh1 | 75 | 0.885 | 0.0397 | No |
| 2 | Pex2 | 230 | 0.730 | 0.0674 | No |
| 3 | Pex5 | 745 | 0.541 | 0.0662 | No |
| 4 | Abcd3 | 802 | 0.527 | 0.0892 | No |
| 5 | Pex11b | 1346 | 0.444 | 0.0816 | No |
| 6 | Fdps | 1608 | 0.412 | 0.0878 | No |
| 7 | Ercc3 | 1779 | 0.393 | 0.0980 | No |
| 8 | Acot8 | 2220 | 0.352 | 0.0914 | No |
| 9 | Sod2 | 2666 | 0.317 | 0.0828 | No |
| 10 | Ech1 | 2722 | 0.314 | 0.0953 | No |
| 11 | Mvp | 2749 | 0.312 | 0.1093 | No |
| 12 | Dhcr24 | 3037 | 0.295 | 0.1083 | No |
| 13 | Sema3c | 3063 | 0.293 | 0.1214 | No |
| 14 | Cadm1 | 3077 | 0.293 | 0.1352 | No |
| 15 | Pex11a | 3131 | 0.289 | 0.1467 | No |
| 16 | Pex13 | 3419 | 0.269 | 0.1443 | No |
| 17 | Sult2b1 | 3552 | 0.261 | 0.1500 | No |
| 18 | Slc23a2 | 3872 | 0.244 | 0.1447 | No |
| 19 | Vps4b | 3956 | 0.238 | 0.1519 | No |
| 20 | Acsl1 | 4068 | 0.231 | 0.1573 | No |
| 21 | Msh2 | 4091 | 0.230 | 0.1675 | No |
| 22 | Dhrs3 | 4370 | 0.213 | 0.1629 | No |
| 23 | Retsat | 4548 | 0.206 | 0.1634 | No |
| 24 | Abcd2 | 4736 | 0.195 | 0.1629 | No |
| 25 | Acsl5 | 5040 | 0.181 | 0.1553 | No |
| 26 | Cdk7 | 5262 | 0.170 | 0.1517 | No |
| 27 | Gnpat | 5504 | 0.158 | 0.1463 | No |
| 28 | Acaa1a | 5866 | 0.143 | 0.1337 | No |
| 29 | Prdx1 | 6072 | 0.133 | 0.1291 | No |
| 30 | Siah1a | 6495 | 0.115 | 0.1118 | No |
| 31 | Prdx5 | 6674 | 0.108 | 0.1074 | No |
| 32 | Abcc5 | 6793 | 0.102 | 0.1060 | No |
| 33 | Lonp2 | 6994 | 0.094 | 0.0997 | No |
| 34 | Cat | 7365 | 0.082 | 0.0836 | No |
| 35 | Mlycd | 7427 | 0.079 | 0.0842 | No |
| 36 | Abcb9 | 7715 | 0.068 | 0.0719 | No |
| 37 | Top2a | 7792 | 0.065 | 0.0710 | No |
| 38 | Hsd17b4 | 7803 | 0.065 | 0.0736 | No |
| 39 | Acsl4 | 8168 | 0.051 | 0.0563 | No |
| 40 | Abcb4 | 8474 | 0.037 | 0.0415 | No |
| 41 | Crat | 8555 | 0.034 | 0.0388 | No |
| 42 | Cln8 | 8681 | 0.029 | 0.0334 | No |
| 43 | Idh2 | 9138 | 0.012 | 0.0091 | No |
| 44 | Rdh11 | 9772 | -0.009 | -0.0250 | No |
| 45 | Abcd1 | 9901 | -0.013 | -0.0313 | No |
| 46 | Sod1 | 10080 | -0.019 | -0.0401 | No |
| 47 | Ide | 10099 | -0.020 | -0.0401 | No |
| 48 | Ehhadh | 10149 | -0.022 | -0.0417 | No |
| 49 | Ephx2 | 10155 | -0.022 | -0.0409 | No |
| 50 | Cln6 | 10271 | -0.026 | -0.0458 | No |
| 51 | Hsd17b11 | 10891 | -0.050 | -0.0772 | No |
| 52 | Bcl10 | 11012 | -0.053 | -0.0811 | No |
| 53 | Ctbp1 | 11586 | -0.075 | -0.1087 | No |
| 54 | Isoc1 | 12094 | -0.094 | -0.1317 | No |
| 55 | Hsd3b7 | 12145 | -0.096 | -0.1296 | No |
| 56 | Nudt19 | 12199 | -0.098 | -0.1276 | No |
| 57 | Smarcc1 | 12526 | -0.112 | -0.1399 | No |
| 58 | Cnbp | 12891 | -0.127 | -0.1534 | No |
| 59 | Scp2 | 13699 | -0.164 | -0.1894 | No |
| 60 | Fis1 | 13947 | -0.175 | -0.1942 | No |
| 61 | Atxn1 | 14118 | -0.184 | -0.1943 | Yes |
| 62 | Pabpc1 | 14155 | -0.186 | -0.1871 | Yes |
| 63 | Itgb1bp1 | 14213 | -0.189 | -0.1808 | Yes |
| 64 | Idi1 | 14434 | -0.201 | -0.1829 | Yes |
| 65 | Dlg4 | 14725 | -0.215 | -0.1881 | Yes |
| 66 | Slc27a2 | 15004 | -0.231 | -0.1918 | Yes |
| 67 | Hmgcl | 15069 | -0.234 | -0.1837 | Yes |
| 68 | Pex14 | 15354 | -0.250 | -0.1868 | Yes |
| 69 | Elovl5 | 15371 | -0.251 | -0.1752 | Yes |
| 70 | Slc25a4 | 15584 | -0.265 | -0.1737 | Yes |
| 71 | Ercc1 | 15747 | -0.275 | -0.1689 | Yes |
| 72 | Esr2 | 15768 | -0.277 | -0.1563 | Yes |
| 73 | Pex6 | 15820 | -0.281 | -0.1452 | Yes |
| 74 | Slc25a19 | 15852 | -0.283 | -0.1329 | Yes |
| 75 | Aldh1a1 | 16004 | -0.293 | -0.1266 | Yes |
| 76 | Ywhah | 16711 | -0.347 | -0.1480 | Yes |
| 77 | Acox1 | 16891 | -0.362 | -0.1398 | Yes |
| 78 | Slc25a17 | 16899 | -0.363 | -0.1222 | Yes |
| 79 | Eci2 | 16941 | -0.367 | -0.1063 | Yes |
| 80 | Slc35b2 | 17002 | -0.374 | -0.0910 | Yes |
| 81 | Aldh9a1 | 17040 | -0.379 | -0.0743 | Yes |
| 82 | Tspo | 17614 | -0.459 | -0.0828 | Yes |
| 83 | Fads1 | 17643 | -0.465 | -0.0613 | Yes |
| 84 | Gstk1 | 17668 | -0.470 | -0.0393 | Yes |
| 85 | Hras | 17761 | -0.484 | -0.0204 | Yes |
| 86 | Cacna1b | 17890 | -0.514 | -0.0019 | Yes |
| 87 | Abcb1a | 18177 | -0.609 | 0.0126 | Yes |