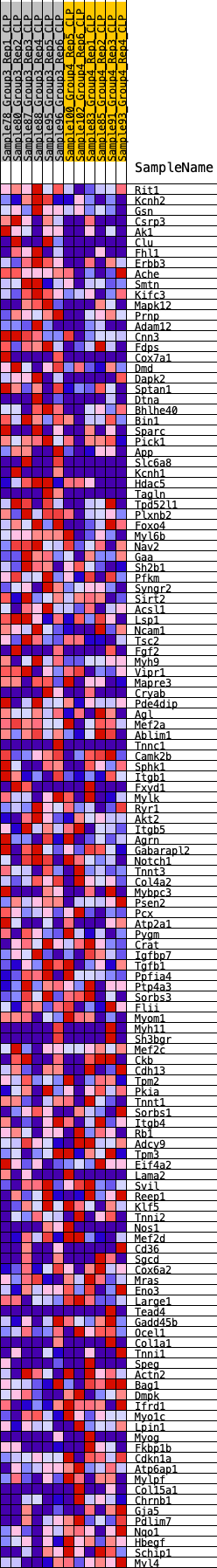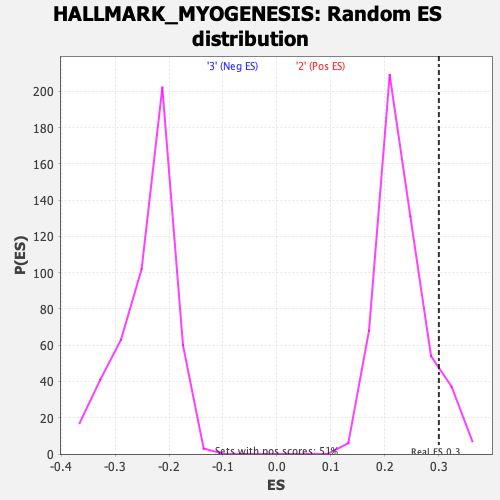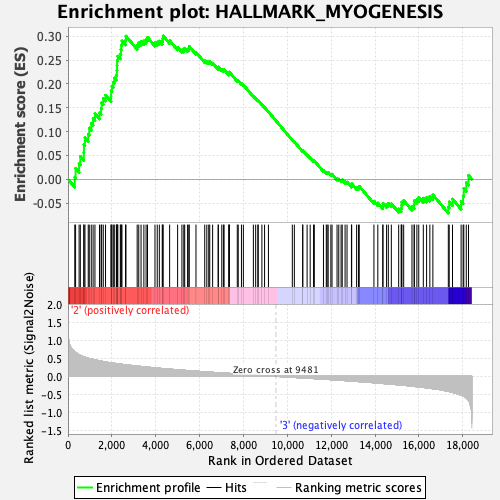
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.30066267 |
| Normalized Enrichment Score (NES) | 1.291532 |
| Nominal p-value | 0.103515625 |
| FDR q-value | 0.5010014 |
| FWER p-Value | 0.817 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rit1 | 309 | 0.680 | 0.0043 | Yes |
| 2 | Kcnh2 | 346 | 0.664 | 0.0230 | Yes |
| 3 | Gsn | 506 | 0.597 | 0.0329 | Yes |
| 4 | Csrp3 | 562 | 0.582 | 0.0480 | Yes |
| 5 | Ak1 | 720 | 0.546 | 0.0564 | Yes |
| 6 | Clu | 727 | 0.545 | 0.0730 | Yes |
| 7 | Fhl1 | 773 | 0.533 | 0.0872 | Yes |
| 8 | Erbb3 | 923 | 0.504 | 0.0947 | Yes |
| 9 | Ache | 969 | 0.494 | 0.1076 | Yes |
| 10 | Smtn | 1060 | 0.485 | 0.1178 | Yes |
| 11 | Kifc3 | 1144 | 0.472 | 0.1279 | Yes |
| 12 | Mapk12 | 1228 | 0.456 | 0.1376 | Yes |
| 13 | Prnp | 1437 | 0.432 | 0.1397 | Yes |
| 14 | Adam12 | 1509 | 0.422 | 0.1489 | Yes |
| 15 | Cnn3 | 1534 | 0.420 | 0.1607 | Yes |
| 16 | Fdps | 1608 | 0.412 | 0.1695 | Yes |
| 17 | Cox7a1 | 1711 | 0.398 | 0.1763 | Yes |
| 18 | Dmd | 1964 | 0.375 | 0.1742 | Yes |
| 19 | Dapk2 | 1967 | 0.374 | 0.1857 | Yes |
| 20 | Sptan1 | 2003 | 0.372 | 0.1954 | Yes |
| 21 | Dtna | 2064 | 0.368 | 0.2036 | Yes |
| 22 | Bhlhe40 | 2110 | 0.363 | 0.2124 | Yes |
| 23 | Bin1 | 2203 | 0.353 | 0.2184 | Yes |
| 24 | Sparc | 2226 | 0.352 | 0.2281 | Yes |
| 25 | Pick1 | 2235 | 0.351 | 0.2386 | Yes |
| 26 | App | 2240 | 0.350 | 0.2493 | Yes |
| 27 | Slc6a8 | 2271 | 0.346 | 0.2584 | Yes |
| 28 | Kcnh1 | 2384 | 0.338 | 0.2628 | Yes |
| 29 | Hdac5 | 2411 | 0.337 | 0.2719 | Yes |
| 30 | Tagln | 2428 | 0.336 | 0.2815 | Yes |
| 31 | Tpd52l1 | 2459 | 0.335 | 0.2902 | Yes |
| 32 | Plxnb2 | 2623 | 0.321 | 0.2913 | Yes |
| 33 | Foxo4 | 2641 | 0.319 | 0.3003 | Yes |
| 34 | Myl6b | 3153 | 0.288 | 0.2813 | Yes |
| 35 | Nav2 | 3225 | 0.283 | 0.2862 | Yes |
| 36 | Gaa | 3330 | 0.275 | 0.2891 | Yes |
| 37 | Sh2b1 | 3459 | 0.267 | 0.2904 | Yes |
| 38 | Pfkm | 3560 | 0.260 | 0.2930 | Yes |
| 39 | Syngr2 | 3624 | 0.256 | 0.2976 | Yes |
| 40 | Sirt2 | 3965 | 0.238 | 0.2863 | Yes |
| 41 | Acsl1 | 4068 | 0.231 | 0.2880 | Yes |
| 42 | Lsp1 | 4163 | 0.226 | 0.2899 | Yes |
| 43 | Ncam1 | 4296 | 0.217 | 0.2894 | Yes |
| 44 | Tsc2 | 4316 | 0.216 | 0.2951 | Yes |
| 45 | Fgf2 | 4337 | 0.215 | 0.3007 | Yes |
| 46 | Myh9 | 4633 | 0.201 | 0.2908 | No |
| 47 | Vipr1 | 4997 | 0.183 | 0.2766 | No |
| 48 | Mapre3 | 5193 | 0.174 | 0.2714 | No |
| 49 | Cryab | 5279 | 0.169 | 0.2720 | No |
| 50 | Pde4dip | 5320 | 0.167 | 0.2750 | No |
| 51 | Agl | 5451 | 0.161 | 0.2729 | No |
| 52 | Mef2a | 5510 | 0.157 | 0.2746 | No |
| 53 | Ablim1 | 5524 | 0.157 | 0.2788 | No |
| 54 | Tnnc1 | 5834 | 0.144 | 0.2664 | No |
| 55 | Camk2b | 6233 | 0.127 | 0.2485 | No |
| 56 | Sphk1 | 6328 | 0.122 | 0.2472 | No |
| 57 | Itgb1 | 6416 | 0.119 | 0.2461 | No |
| 58 | Fxyd1 | 6461 | 0.117 | 0.2474 | No |
| 59 | Mylk | 6593 | 0.111 | 0.2437 | No |
| 60 | Ryr1 | 6844 | 0.099 | 0.2331 | No |
| 61 | Akt2 | 6855 | 0.099 | 0.2356 | No |
| 62 | Itgb5 | 7010 | 0.092 | 0.2301 | No |
| 63 | Agrn | 7084 | 0.090 | 0.2289 | No |
| 64 | Gabarapl2 | 7115 | 0.088 | 0.2300 | No |
| 65 | Notch1 | 7327 | 0.084 | 0.2210 | No |
| 66 | Tnnt3 | 7351 | 0.083 | 0.2223 | No |
| 67 | Col4a2 | 7352 | 0.083 | 0.2249 | No |
| 68 | Mybpc3 | 7717 | 0.068 | 0.2071 | No |
| 69 | Psen2 | 7760 | 0.066 | 0.2069 | No |
| 70 | Pcx | 7907 | 0.061 | 0.2008 | No |
| 71 | Atp2a1 | 7994 | 0.058 | 0.1979 | No |
| 72 | Pygm | 8450 | 0.038 | 0.1742 | No |
| 73 | Crat | 8555 | 0.034 | 0.1695 | No |
| 74 | Igfbp7 | 8655 | 0.030 | 0.1650 | No |
| 75 | Tgfb1 | 8671 | 0.029 | 0.1651 | No |
| 76 | Ppfia4 | 8839 | 0.023 | 0.1567 | No |
| 77 | Ptp4a3 | 8961 | 0.019 | 0.1507 | No |
| 78 | Sorbs3 | 9137 | 0.012 | 0.1415 | No |
| 79 | Flii | 10226 | -0.024 | 0.0827 | No |
| 80 | Myom1 | 10318 | -0.028 | 0.0786 | No |
| 81 | Myh11 | 10699 | -0.043 | 0.0591 | No |
| 82 | Sh3bgr | 10702 | -0.043 | 0.0603 | No |
| 83 | Mef2c | 10900 | -0.050 | 0.0511 | No |
| 84 | Ckb | 11040 | -0.054 | 0.0452 | No |
| 85 | Cdh13 | 11193 | -0.060 | 0.0387 | No |
| 86 | Tpm2 | 11228 | -0.061 | 0.0388 | No |
| 87 | Pkia | 11646 | -0.077 | 0.0183 | No |
| 88 | Tnnt1 | 11774 | -0.082 | 0.0139 | No |
| 89 | Sorbs1 | 11812 | -0.083 | 0.0145 | No |
| 90 | Itgb4 | 11860 | -0.085 | 0.0146 | No |
| 91 | Rb1 | 11982 | -0.090 | 0.0108 | No |
| 92 | Adcy9 | 12036 | -0.092 | 0.0107 | No |
| 93 | Tpm3 | 12265 | -0.101 | 0.0014 | No |
| 94 | Eif4a2 | 12343 | -0.104 | 0.0004 | No |
| 95 | Lama2 | 12449 | -0.109 | -0.0019 | No |
| 96 | Svil | 12501 | -0.111 | -0.0013 | No |
| 97 | Reep1 | 12631 | -0.116 | -0.0047 | No |
| 98 | Klf5 | 12720 | -0.120 | -0.0058 | No |
| 99 | Tnni2 | 12921 | -0.129 | -0.0127 | No |
| 100 | Nos1 | 12932 | -0.130 | -0.0092 | No |
| 101 | Mef2d | 13157 | -0.139 | -0.0171 | No |
| 102 | Cd36 | 13241 | -0.143 | -0.0172 | No |
| 103 | Sgcd | 13276 | -0.144 | -0.0146 | No |
| 104 | Cox6a2 | 13946 | -0.175 | -0.0457 | No |
| 105 | Mras | 14123 | -0.185 | -0.0496 | No |
| 106 | Eno3 | 14350 | -0.196 | -0.0559 | No |
| 107 | Large1 | 14357 | -0.197 | -0.0501 | No |
| 108 | Tead4 | 14523 | -0.205 | -0.0527 | No |
| 109 | Gadd45b | 14602 | -0.207 | -0.0505 | No |
| 110 | Ocel1 | 14739 | -0.215 | -0.0513 | No |
| 111 | Col1a1 | 15074 | -0.235 | -0.0622 | No |
| 112 | Tnni1 | 15187 | -0.239 | -0.0609 | No |
| 113 | Speg | 15208 | -0.240 | -0.0546 | No |
| 114 | Actn2 | 15219 | -0.241 | -0.0476 | No |
| 115 | Bag1 | 15302 | -0.246 | -0.0444 | No |
| 116 | Dmpk | 15681 | -0.272 | -0.0566 | No |
| 117 | Ifrd1 | 15782 | -0.278 | -0.0535 | No |
| 118 | Myo1c | 15784 | -0.278 | -0.0449 | No |
| 119 | Lpin1 | 15904 | -0.287 | -0.0424 | No |
| 120 | Myog | 15989 | -0.293 | -0.0379 | No |
| 121 | Fkbp1b | 16197 | -0.307 | -0.0397 | No |
| 122 | Cdkn1a | 16347 | -0.318 | -0.0379 | No |
| 123 | Atp6ap1 | 16495 | -0.330 | -0.0357 | No |
| 124 | Mylpf | 16633 | -0.341 | -0.0326 | No |
| 125 | Col15a1 | 17337 | -0.416 | -0.0581 | No |
| 126 | Chrnb1 | 17378 | -0.421 | -0.0472 | No |
| 127 | Gja5 | 17528 | -0.445 | -0.0415 | No |
| 128 | Pdlim7 | 17920 | -0.519 | -0.0468 | No |
| 129 | Nqo1 | 18011 | -0.541 | -0.0349 | No |
| 130 | Hbegf | 18043 | -0.551 | -0.0194 | No |
| 131 | Schip1 | 18156 | -0.598 | -0.0069 | No |
| 132 | Myl4 | 18257 | -0.663 | 0.0083 | No |