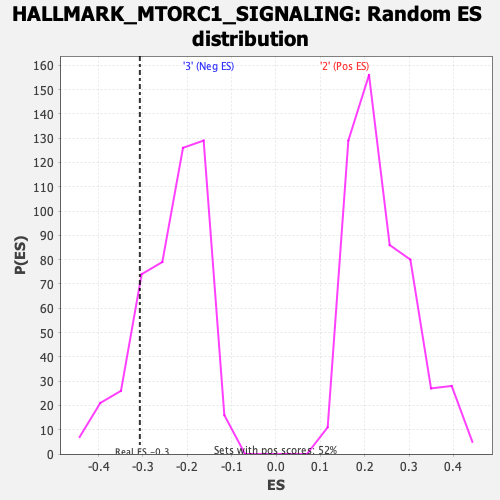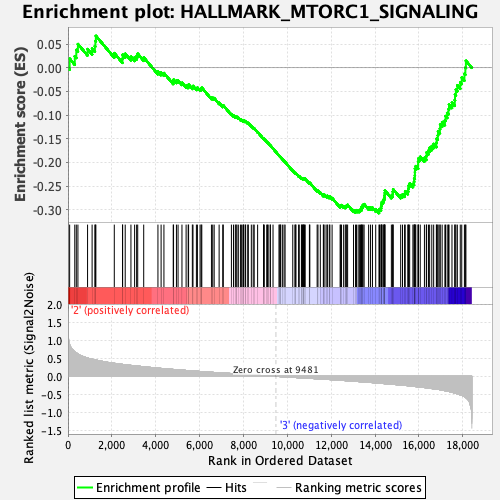
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_MTORC1_SIGNALING |
| Enrichment Score (ES) | -0.30709177 |
| Normalized Enrichment Score (NES) | -1.2981935 |
| Nominal p-value | 0.17782427 |
| FDR q-value | 0.55142903 |
| FWER p-Value | 0.791 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idh1 | 75 | 0.885 | 0.0192 | No |
| 2 | Rit1 | 309 | 0.680 | 0.0243 | No |
| 3 | Serpinh1 | 378 | 0.651 | 0.0378 | No |
| 4 | Gla | 452 | 0.618 | 0.0501 | No |
| 5 | Got1 | 892 | 0.512 | 0.0395 | No |
| 6 | Psph | 1099 | 0.478 | 0.0408 | No |
| 7 | Tm7sf2 | 1226 | 0.457 | 0.0459 | No |
| 8 | Ldlr | 1248 | 0.454 | 0.0567 | No |
| 9 | Hmgcs1 | 1270 | 0.451 | 0.0674 | No |
| 10 | Bhlhe40 | 2110 | 0.363 | 0.0309 | No |
| 11 | Gbe1 | 2484 | 0.332 | 0.0192 | No |
| 12 | Sqstm1 | 2491 | 0.332 | 0.0276 | No |
| 13 | Slc6a6 | 2608 | 0.322 | 0.0297 | No |
| 14 | Pik3r3 | 2869 | 0.305 | 0.0235 | No |
| 15 | Dhcr24 | 3037 | 0.295 | 0.0221 | No |
| 16 | Niban1 | 3129 | 0.289 | 0.0247 | No |
| 17 | Cfp | 3178 | 0.287 | 0.0297 | No |
| 18 | Psmb5 | 3451 | 0.267 | 0.0218 | No |
| 19 | Pitpnb | 4100 | 0.229 | -0.0078 | No |
| 20 | Mthfd2l | 4244 | 0.221 | -0.0098 | No |
| 21 | Ak4 | 4373 | 0.213 | -0.0112 | No |
| 22 | Xbp1 | 4797 | 0.192 | -0.0293 | No |
| 23 | Ccnf | 4811 | 0.191 | -0.0250 | No |
| 24 | Atp2a2 | 4943 | 0.185 | -0.0273 | No |
| 25 | Tfrc | 5017 | 0.182 | -0.0265 | No |
| 26 | Lgmn | 5195 | 0.173 | -0.0317 | No |
| 27 | Sytl2 | 5384 | 0.164 | -0.0377 | No |
| 28 | G6pdx | 5479 | 0.159 | -0.0387 | No |
| 29 | Ddit4 | 5497 | 0.158 | -0.0354 | No |
| 30 | Actr3 | 5668 | 0.150 | -0.0408 | No |
| 31 | Dapp1 | 5697 | 0.149 | -0.0384 | No |
| 32 | Plk1 | 5853 | 0.144 | -0.0431 | No |
| 33 | Skap2 | 5899 | 0.141 | -0.0419 | No |
| 34 | Vldlr | 6020 | 0.136 | -0.0449 | No |
| 35 | Prdx1 | 6072 | 0.133 | -0.0442 | No |
| 36 | Slc7a5 | 6099 | 0.132 | -0.0421 | No |
| 37 | Add3 | 6539 | 0.113 | -0.0632 | No |
| 38 | Bub1 | 6597 | 0.111 | -0.0634 | No |
| 39 | Sc5d | 6663 | 0.108 | -0.0642 | No |
| 40 | Cops5 | 6889 | 0.098 | -0.0739 | No |
| 41 | Slc2a3 | 7055 | 0.091 | -0.0806 | No |
| 42 | Pno1 | 7078 | 0.090 | -0.0794 | No |
| 43 | Gpi1 | 7449 | 0.078 | -0.0977 | No |
| 44 | Adipor2 | 7544 | 0.075 | -0.1009 | No |
| 45 | Slc1a5 | 7627 | 0.071 | -0.1035 | No |
| 46 | Psmd14 | 7661 | 0.070 | -0.1035 | No |
| 47 | Ccng1 | 7700 | 0.069 | -0.1037 | No |
| 48 | Insig1 | 7764 | 0.066 | -0.1055 | No |
| 49 | Ctsc | 7860 | 0.063 | -0.1090 | No |
| 50 | Cyb5b | 7903 | 0.061 | -0.1097 | No |
| 51 | Mcm4 | 7972 | 0.059 | -0.1119 | No |
| 52 | Cd9 | 8003 | 0.058 | -0.1120 | No |
| 53 | Elovl6 | 8010 | 0.058 | -0.1108 | No |
| 54 | Gsr | 8081 | 0.055 | -0.1132 | No |
| 55 | Hspa4 | 8098 | 0.054 | -0.1127 | No |
| 56 | Atp6v1d | 8195 | 0.050 | -0.1166 | No |
| 57 | Psmc6 | 8219 | 0.049 | -0.1166 | No |
| 58 | Bcat1 | 8357 | 0.043 | -0.1230 | No |
| 59 | Pnp | 8442 | 0.039 | -0.1266 | No |
| 60 | Gmps | 8488 | 0.037 | -0.1281 | No |
| 61 | Psma3 | 8644 | 0.030 | -0.1358 | No |
| 62 | Calr | 8913 | 0.020 | -0.1499 | No |
| 63 | Hspa9 | 8914 | 0.020 | -0.1494 | No |
| 64 | Hk2 | 8948 | 0.019 | -0.1507 | No |
| 65 | Sqle | 9059 | 0.015 | -0.1564 | No |
| 66 | Aurka | 9096 | 0.013 | -0.1580 | No |
| 67 | Me1 | 9124 | 0.012 | -0.1592 | No |
| 68 | Rpn1 | 9220 | 0.009 | -0.1641 | No |
| 69 | Sla | 9222 | 0.009 | -0.1640 | No |
| 70 | Stard4 | 9346 | 0.004 | -0.1706 | No |
| 71 | Aldoa | 9618 | -0.003 | -0.1854 | No |
| 72 | Psme3 | 9656 | -0.004 | -0.1873 | No |
| 73 | Tpi1 | 9673 | -0.005 | -0.1881 | No |
| 74 | Ufm1 | 9695 | -0.006 | -0.1890 | No |
| 75 | Rdh11 | 9772 | -0.009 | -0.1930 | No |
| 76 | Mthfd2 | 9848 | -0.012 | -0.1968 | No |
| 77 | Hmgcr | 9896 | -0.013 | -0.1990 | No |
| 78 | Actr2 | 10246 | -0.025 | -0.2175 | No |
| 79 | Slc1a4 | 10342 | -0.029 | -0.2220 | No |
| 80 | Lta4h | 10380 | -0.031 | -0.2232 | No |
| 81 | Fdxr | 10386 | -0.031 | -0.2226 | No |
| 82 | Ube2d3 | 10512 | -0.036 | -0.2285 | No |
| 83 | Tomm40 | 10541 | -0.037 | -0.2291 | No |
| 84 | Abcf2 | 10640 | -0.040 | -0.2334 | No |
| 85 | Ifi30 | 10660 | -0.041 | -0.2334 | No |
| 86 | Ero1a | 10689 | -0.042 | -0.2338 | No |
| 87 | Gsk3b | 10721 | -0.043 | -0.2344 | No |
| 88 | Nup205 | 10732 | -0.043 | -0.2338 | No |
| 89 | Ppp1r15a | 10749 | -0.044 | -0.2335 | No |
| 90 | Cth | 10781 | -0.046 | -0.2340 | No |
| 91 | M6pr | 10813 | -0.047 | -0.2344 | No |
| 92 | Ung | 11005 | -0.053 | -0.2435 | No |
| 93 | Fgl2 | 11024 | -0.054 | -0.2431 | No |
| 94 | Itgb2 | 11357 | -0.066 | -0.2596 | No |
| 95 | Nfil3 | 11395 | -0.067 | -0.2599 | No |
| 96 | Mcm2 | 11502 | -0.071 | -0.2638 | No |
| 97 | Etf1 | 11640 | -0.077 | -0.2693 | No |
| 98 | Rab1a | 11653 | -0.077 | -0.2679 | No |
| 99 | Eno1b | 11713 | -0.079 | -0.2691 | No |
| 100 | Nfyc | 11803 | -0.083 | -0.2718 | No |
| 101 | Slc37a4 | 11822 | -0.084 | -0.2706 | No |
| 102 | Immt | 11910 | -0.087 | -0.2731 | No |
| 103 | Psmg1 | 11938 | -0.088 | -0.2722 | No |
| 104 | Cacybp | 12039 | -0.092 | -0.2753 | No |
| 105 | Stip1 | 12405 | -0.107 | -0.2925 | No |
| 106 | Glrx | 12428 | -0.108 | -0.2908 | No |
| 107 | Ddx39a | 12469 | -0.110 | -0.2902 | No |
| 108 | Tuba4a | 12565 | -0.113 | -0.2924 | No |
| 109 | Srd5a1 | 12640 | -0.117 | -0.2934 | No |
| 110 | Gga2 | 12675 | -0.118 | -0.2921 | No |
| 111 | Hmbs | 12706 | -0.119 | -0.2906 | No |
| 112 | Asns | 12747 | -0.122 | -0.2896 | No |
| 113 | Pdk1 | 13016 | -0.133 | -0.3008 | No |
| 114 | Wars1 | 13108 | -0.137 | -0.3022 | Yes |
| 115 | Tes | 13153 | -0.139 | -0.3009 | Yes |
| 116 | Ppia | 13228 | -0.143 | -0.3012 | Yes |
| 117 | Fkbp2 | 13294 | -0.145 | -0.3010 | Yes |
| 118 | Mllt11 | 13332 | -0.147 | -0.2991 | Yes |
| 119 | Tcea1 | 13380 | -0.149 | -0.2978 | Yes |
| 120 | Gclc | 13382 | -0.149 | -0.2940 | Yes |
| 121 | Gtf2h1 | 13430 | -0.151 | -0.2926 | Yes |
| 122 | Nufip1 | 13447 | -0.151 | -0.2895 | Yes |
| 123 | Hspa5 | 13498 | -0.154 | -0.2881 | Yes |
| 124 | Nfkbib | 13705 | -0.165 | -0.2951 | Yes |
| 125 | Cxcr4 | 13792 | -0.168 | -0.2954 | Yes |
| 126 | Tubg1 | 13879 | -0.172 | -0.2956 | Yes |
| 127 | Dhcr7 | 14027 | -0.179 | -0.2989 | Yes |
| 128 | Trib3 | 14177 | -0.187 | -0.3022 | Yes |
| 129 | Ddit3 | 14199 | -0.189 | -0.2983 | Yes |
| 130 | Acaca | 14268 | -0.192 | -0.2970 | Yes |
| 131 | Polr3g | 14277 | -0.193 | -0.2924 | Yes |
| 132 | Slc2a1 | 14292 | -0.194 | -0.2880 | Yes |
| 133 | Uso1 | 14308 | -0.194 | -0.2837 | Yes |
| 134 | Sec11a | 14362 | -0.197 | -0.2814 | Yes |
| 135 | Hsp90b1 | 14394 | -0.199 | -0.2779 | Yes |
| 136 | Arpc5l | 14419 | -0.200 | -0.2739 | Yes |
| 137 | Idi1 | 14434 | -0.201 | -0.2694 | Yes |
| 138 | Cct6a | 14441 | -0.201 | -0.2645 | Yes |
| 139 | Nmt1 | 14447 | -0.201 | -0.2594 | Yes |
| 140 | Map2k3 | 14741 | -0.215 | -0.2698 | Yes |
| 141 | Pgm1 | 14800 | -0.218 | -0.2673 | Yes |
| 142 | Tmem97 | 14806 | -0.219 | -0.2618 | Yes |
| 143 | Rrm2 | 14823 | -0.220 | -0.2569 | Yes |
| 144 | Psmc2 | 15166 | -0.238 | -0.2694 | Yes |
| 145 | P4ha1 | 15249 | -0.243 | -0.2675 | Yes |
| 146 | Ebp | 15344 | -0.249 | -0.2661 | Yes |
| 147 | Elovl5 | 15371 | -0.251 | -0.2609 | Yes |
| 148 | Hspd1 | 15491 | -0.258 | -0.2606 | Yes |
| 149 | Sord | 15515 | -0.260 | -0.2550 | Yes |
| 150 | Pfkl | 15530 | -0.261 | -0.2489 | Yes |
| 151 | Btg2 | 15575 | -0.265 | -0.2443 | Yes |
| 152 | Canx | 15723 | -0.274 | -0.2452 | Yes |
| 153 | Psmd12 | 15775 | -0.277 | -0.2407 | Yes |
| 154 | Ifrd1 | 15782 | -0.278 | -0.2337 | Yes |
| 155 | Txnrd1 | 15810 | -0.280 | -0.2278 | Yes |
| 156 | Ppa1 | 15814 | -0.280 | -0.2206 | Yes |
| 157 | Acly | 15817 | -0.281 | -0.2133 | Yes |
| 158 | Phgdh | 15854 | -0.283 | -0.2078 | Yes |
| 159 | Uchl5 | 15960 | -0.291 | -0.2059 | Yes |
| 160 | Acsl3 | 15961 | -0.291 | -0.1982 | Yes |
| 161 | Eif2s2 | 15978 | -0.292 | -0.1914 | Yes |
| 162 | Qdpr | 16057 | -0.297 | -0.1878 | Yes |
| 163 | Sdf2l1 | 16248 | -0.310 | -0.1901 | Yes |
| 164 | Psma4 | 16334 | -0.316 | -0.1864 | Yes |
| 165 | Cdkn1a | 16347 | -0.318 | -0.1787 | Yes |
| 166 | Shmt2 | 16432 | -0.325 | -0.1748 | Yes |
| 167 | Tbk1 | 16486 | -0.329 | -0.1690 | Yes |
| 168 | Fads2 | 16584 | -0.338 | -0.1654 | Yes |
| 169 | Ykt6 | 16669 | -0.344 | -0.1610 | Yes |
| 170 | Ssr1 | 16792 | -0.354 | -0.1583 | Yes |
| 171 | Eef1e1 | 16805 | -0.355 | -0.1496 | Yes |
| 172 | Ldha | 16857 | -0.359 | -0.1430 | Yes |
| 173 | Cyp51 | 16869 | -0.360 | -0.1341 | Yes |
| 174 | Plod2 | 16953 | -0.368 | -0.1289 | Yes |
| 175 | Nampt | 16965 | -0.369 | -0.1198 | Yes |
| 176 | Egln3 | 17056 | -0.381 | -0.1147 | Yes |
| 177 | Dhfr | 17181 | -0.396 | -0.1111 | Yes |
| 178 | Pdap1 | 17210 | -0.400 | -0.1020 | Yes |
| 179 | Psmc4 | 17287 | -0.410 | -0.0954 | Yes |
| 180 | Serp1 | 17346 | -0.418 | -0.0876 | Yes |
| 181 | Rpa1 | 17369 | -0.420 | -0.0777 | Yes |
| 182 | Coro1a | 17504 | -0.441 | -0.0735 | Yes |
| 183 | Rrp9 | 17631 | -0.462 | -0.0682 | Yes |
| 184 | Fads1 | 17643 | -0.465 | -0.0565 | Yes |
| 185 | Pgk1 | 17683 | -0.472 | -0.0462 | Yes |
| 186 | Psmd13 | 17748 | -0.481 | -0.0371 | Yes |
| 187 | Slc7a11 | 17882 | -0.513 | -0.0309 | Yes |
| 188 | Edem1 | 17949 | -0.526 | -0.0206 | Yes |
| 189 | Hspe1 | 18071 | -0.562 | -0.0125 | Yes |
| 190 | Cdc25a | 18114 | -0.579 | 0.0005 | Yes |
| 191 | Psat1 | 18139 | -0.592 | 0.0148 | Yes |