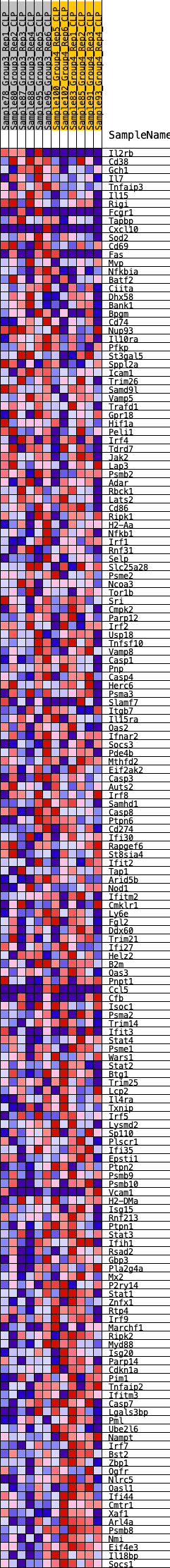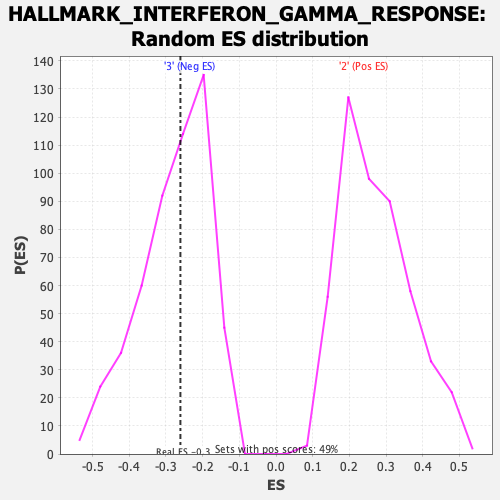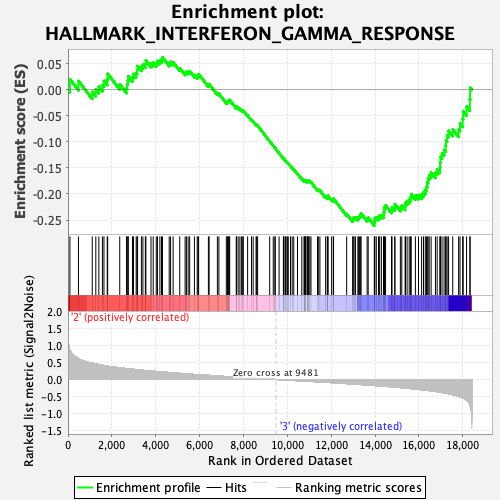
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_INTERFERON_GAMMA_RESPONSE |
| Enrichment Score (ES) | -0.26067558 |
| Normalized Enrichment Score (NES) | -0.93658584 |
| Nominal p-value | 0.48727983 |
| FDR q-value | 0.9327335 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Il2rb | 87 | 0.862 | 0.0203 | No |
| 2 | Cd38 | 476 | 0.610 | 0.0168 | No |
| 3 | Gch1 | 1105 | 0.477 | -0.0037 | No |
| 4 | Il7 | 1268 | 0.451 | 0.0006 | No |
| 5 | Tnfaip3 | 1406 | 0.437 | 0.0058 | No |
| 6 | Il15 | 1569 | 0.416 | 0.0090 | No |
| 7 | Rigi | 1639 | 0.407 | 0.0171 | No |
| 8 | Fcgr1 | 1788 | 0.392 | 0.0204 | No |
| 9 | Tapbp | 1809 | 0.390 | 0.0307 | No |
| 10 | Cxcl10 | 2359 | 0.339 | 0.0104 | No |
| 11 | Sod2 | 2666 | 0.317 | 0.0029 | No |
| 12 | Cd69 | 2692 | 0.316 | 0.0107 | No |
| 13 | Fas | 2729 | 0.313 | 0.0179 | No |
| 14 | Mvp | 2749 | 0.312 | 0.0259 | No |
| 15 | Nfkbia | 2939 | 0.298 | 0.0242 | No |
| 16 | Batf2 | 2987 | 0.296 | 0.0303 | No |
| 17 | Ciita | 3108 | 0.291 | 0.0322 | No |
| 18 | Dhx58 | 3151 | 0.288 | 0.0383 | No |
| 19 | Bank1 | 3172 | 0.287 | 0.0455 | No |
| 20 | Bpgm | 3346 | 0.274 | 0.0440 | No |
| 21 | Cd74 | 3415 | 0.269 | 0.0481 | No |
| 22 | Nup93 | 3526 | 0.263 | 0.0498 | No |
| 23 | Il10ra | 3551 | 0.261 | 0.0561 | No |
| 24 | Pfkp | 3784 | 0.247 | 0.0505 | No |
| 25 | St3gal5 | 3883 | 0.243 | 0.0522 | No |
| 26 | Sppl2a | 4023 | 0.235 | 0.0515 | No |
| 27 | Icam1 | 4078 | 0.231 | 0.0552 | No |
| 28 | Trim26 | 4188 | 0.224 | 0.0558 | No |
| 29 | Samd9l | 4262 | 0.219 | 0.0582 | No |
| 30 | Vamp5 | 4306 | 0.216 | 0.0621 | No |
| 31 | Trafd1 | 4613 | 0.202 | 0.0512 | No |
| 32 | Gpr18 | 4676 | 0.199 | 0.0536 | No |
| 33 | Hif1a | 4792 | 0.193 | 0.0529 | No |
| 34 | Peli1 | 5094 | 0.178 | 0.0416 | No |
| 35 | Irf4 | 5339 | 0.166 | 0.0330 | No |
| 36 | Tdrd7 | 5398 | 0.163 | 0.0346 | No |
| 37 | Jak2 | 5477 | 0.159 | 0.0350 | No |
| 38 | Lap3 | 5546 | 0.156 | 0.0358 | No |
| 39 | Psmb2 | 5763 | 0.146 | 0.0282 | No |
| 40 | Adar | 5891 | 0.142 | 0.0254 | No |
| 41 | Rbck1 | 5930 | 0.140 | 0.0274 | No |
| 42 | Lats2 | 5955 | 0.139 | 0.0301 | No |
| 43 | Cd86 | 6400 | 0.120 | 0.0092 | No |
| 44 | Ripk1 | 6433 | 0.118 | 0.0109 | No |
| 45 | H2-Aa | 6810 | 0.101 | -0.0067 | No |
| 46 | Nfkb1 | 6878 | 0.098 | -0.0076 | No |
| 47 | Irf1 | 7217 | 0.087 | -0.0236 | No |
| 48 | Rnf31 | 7273 | 0.086 | -0.0241 | No |
| 49 | Selp | 7295 | 0.085 | -0.0227 | No |
| 50 | Slc25a28 | 7306 | 0.085 | -0.0208 | No |
| 51 | Psme2 | 7348 | 0.083 | -0.0207 | No |
| 52 | Ncoa3 | 7371 | 0.082 | -0.0195 | No |
| 53 | Tor1b | 7670 | 0.070 | -0.0338 | No |
| 54 | Sri | 7685 | 0.069 | -0.0326 | No |
| 55 | Cmpk2 | 7756 | 0.066 | -0.0345 | No |
| 56 | Parp12 | 7835 | 0.064 | -0.0369 | No |
| 57 | Irf2 | 7904 | 0.061 | -0.0388 | No |
| 58 | Usp18 | 7963 | 0.060 | -0.0403 | No |
| 59 | Tnfsf10 | 7991 | 0.059 | -0.0401 | No |
| 60 | Vamp8 | 8188 | 0.050 | -0.0493 | No |
| 61 | Casp1 | 8365 | 0.042 | -0.0578 | No |
| 62 | Pnp | 8442 | 0.039 | -0.0608 | No |
| 63 | Casp4 | 8572 | 0.033 | -0.0669 | No |
| 64 | Herc6 | 8600 | 0.032 | -0.0674 | No |
| 65 | Psma3 | 8644 | 0.030 | -0.0689 | No |
| 66 | Slamf7 | 9194 | 0.010 | -0.0987 | No |
| 67 | Itgb7 | 9356 | 0.004 | -0.1074 | No |
| 68 | Il15ra | 9431 | 0.002 | -0.1114 | No |
| 69 | Oas2 | 9449 | 0.001 | -0.1123 | No |
| 70 | Ifnar2 | 9622 | -0.003 | -0.1217 | No |
| 71 | Socs3 | 9818 | -0.011 | -0.1321 | No |
| 72 | Pde4b | 9829 | -0.011 | -0.1323 | No |
| 73 | Mthfd2 | 9848 | -0.012 | -0.1329 | No |
| 74 | Eif2ak2 | 9919 | -0.014 | -0.1364 | No |
| 75 | Casp3 | 9943 | -0.015 | -0.1372 | No |
| 76 | Auts2 | 10010 | -0.017 | -0.1403 | No |
| 77 | Irf8 | 10040 | -0.018 | -0.1414 | No |
| 78 | Samhd1 | 10144 | -0.022 | -0.1464 | No |
| 79 | Casp8 | 10219 | -0.024 | -0.1498 | No |
| 80 | Ptpn6 | 10291 | -0.027 | -0.1529 | No |
| 81 | Cd274 | 10460 | -0.034 | -0.1611 | No |
| 82 | Ifi30 | 10660 | -0.041 | -0.1708 | No |
| 83 | Rapgef6 | 10759 | -0.045 | -0.1748 | No |
| 84 | St8sia4 | 10791 | -0.046 | -0.1752 | No |
| 85 | Ifit2 | 10796 | -0.046 | -0.1741 | No |
| 86 | Tap1 | 10831 | -0.047 | -0.1746 | No |
| 87 | Arid5b | 10847 | -0.048 | -0.1740 | No |
| 88 | Nod1 | 10914 | -0.050 | -0.1761 | No |
| 89 | Ifitm2 | 10930 | -0.051 | -0.1755 | No |
| 90 | Cmklr1 | 10942 | -0.051 | -0.1746 | No |
| 91 | Ly6e | 10973 | -0.052 | -0.1747 | No |
| 92 | Fgl2 | 11024 | -0.054 | -0.1759 | No |
| 93 | Ddx60 | 11076 | -0.055 | -0.1771 | No |
| 94 | Trim21 | 11374 | -0.067 | -0.1914 | No |
| 95 | Ifi27 | 11410 | -0.068 | -0.1914 | No |
| 96 | Helz2 | 11483 | -0.070 | -0.1933 | No |
| 97 | B2m | 11744 | -0.081 | -0.2052 | No |
| 98 | Oas3 | 11819 | -0.084 | -0.2068 | No |
| 99 | Pnpt1 | 11847 | -0.084 | -0.2058 | No |
| 100 | Ccl5 | 11848 | -0.084 | -0.2034 | No |
| 101 | Cfb | 12018 | -0.092 | -0.2100 | No |
| 102 | Isoc1 | 12094 | -0.094 | -0.2113 | No |
| 103 | Psma2 | 12106 | -0.095 | -0.2092 | No |
| 104 | Trim14 | 12703 | -0.119 | -0.2384 | No |
| 105 | Ifit3 | 12975 | -0.131 | -0.2494 | No |
| 106 | Stat4 | 13000 | -0.132 | -0.2469 | No |
| 107 | Psme1 | 13043 | -0.134 | -0.2453 | No |
| 108 | Wars1 | 13108 | -0.137 | -0.2448 | No |
| 109 | Stat2 | 13215 | -0.142 | -0.2465 | No |
| 110 | Btg1 | 13249 | -0.143 | -0.2441 | No |
| 111 | Trim25 | 13293 | -0.145 | -0.2422 | No |
| 112 | Lcp2 | 13342 | -0.147 | -0.2406 | No |
| 113 | Il4ra | 13358 | -0.148 | -0.2371 | No |
| 114 | Txnip | 13636 | -0.161 | -0.2476 | No |
| 115 | Irf5 | 13684 | -0.163 | -0.2454 | No |
| 116 | Lysmd2 | 13963 | -0.176 | -0.2555 | Yes |
| 117 | Sp110 | 13976 | -0.177 | -0.2511 | Yes |
| 118 | Plscr1 | 13986 | -0.177 | -0.2464 | Yes |
| 119 | Ifi35 | 14054 | -0.181 | -0.2448 | Yes |
| 120 | Epsti1 | 14154 | -0.186 | -0.2448 | Yes |
| 121 | Ptpn2 | 14205 | -0.189 | -0.2420 | Yes |
| 122 | Psmb9 | 14285 | -0.193 | -0.2407 | Yes |
| 123 | Psmb10 | 14384 | -0.198 | -0.2403 | Yes |
| 124 | Vcam1 | 14397 | -0.199 | -0.2352 | Yes |
| 125 | H2-DMa | 14411 | -0.200 | -0.2301 | Yes |
| 126 | Isg15 | 14424 | -0.200 | -0.2249 | Yes |
| 127 | Rnf213 | 14476 | -0.203 | -0.2218 | Yes |
| 128 | Ptpn1 | 14747 | -0.215 | -0.2303 | Yes |
| 129 | Stat3 | 14771 | -0.216 | -0.2253 | Yes |
| 130 | Ifih1 | 14897 | -0.224 | -0.2256 | Yes |
| 131 | Rsad2 | 14902 | -0.225 | -0.2193 | Yes |
| 132 | Gbp3 | 15151 | -0.236 | -0.2260 | Yes |
| 133 | Pla2g4a | 15216 | -0.241 | -0.2225 | Yes |
| 134 | Mx2 | 15368 | -0.251 | -0.2235 | Yes |
| 135 | P2ry14 | 15384 | -0.252 | -0.2170 | Yes |
| 136 | Stat1 | 15458 | -0.256 | -0.2135 | Yes |
| 137 | Znfx1 | 15553 | -0.263 | -0.2110 | Yes |
| 138 | Rtp4 | 15610 | -0.267 | -0.2063 | Yes |
| 139 | Irf9 | 15645 | -0.270 | -0.2003 | Yes |
| 140 | Marchf1 | 15836 | -0.282 | -0.2025 | Yes |
| 141 | Ripk2 | 15979 | -0.292 | -0.2018 | Yes |
| 142 | Myd88 | 16120 | -0.303 | -0.2006 | Yes |
| 143 | Isg20 | 16206 | -0.308 | -0.1963 | Yes |
| 144 | Parp14 | 16288 | -0.312 | -0.1917 | Yes |
| 145 | Cdkn1a | 16347 | -0.318 | -0.1856 | Yes |
| 146 | Pim1 | 16374 | -0.320 | -0.1777 | Yes |
| 147 | Tnfaip2 | 16414 | -0.323 | -0.1704 | Yes |
| 148 | Ifitm3 | 16464 | -0.328 | -0.1636 | Yes |
| 149 | Casp7 | 16553 | -0.335 | -0.1586 | Yes |
| 150 | Lgals3bp | 16749 | -0.351 | -0.1591 | Yes |
| 151 | Pml | 16828 | -0.358 | -0.1530 | Yes |
| 152 | Ube2l6 | 16956 | -0.368 | -0.1492 | Yes |
| 153 | Nampt | 16965 | -0.369 | -0.1389 | Yes |
| 154 | Irf7 | 16979 | -0.372 | -0.1288 | Yes |
| 155 | Bst2 | 17051 | -0.380 | -0.1216 | Yes |
| 156 | Zbp1 | 17144 | -0.393 | -0.1152 | Yes |
| 157 | Ogfr | 17215 | -0.402 | -0.1073 | Yes |
| 158 | Nlrc5 | 17231 | -0.404 | -0.0964 | Yes |
| 159 | Oasl1 | 17283 | -0.410 | -0.0873 | Yes |
| 160 | Ifi44 | 17349 | -0.418 | -0.0787 | Yes |
| 161 | Cmtr1 | 17539 | -0.447 | -0.0760 | Yes |
| 162 | Xaf1 | 17807 | -0.494 | -0.0763 | Yes |
| 163 | Arl4a | 17869 | -0.509 | -0.0648 | Yes |
| 164 | Psmb8 | 17992 | -0.536 | -0.0558 | Yes |
| 165 | Nmi | 18013 | -0.541 | -0.0412 | Yes |
| 166 | Eif4e3 | 18169 | -0.605 | -0.0321 | Yes |
| 167 | Il18bp | 18320 | -0.762 | -0.0181 | Yes |
| 168 | Socs1 | 18332 | -0.785 | 0.0042 | Yes |