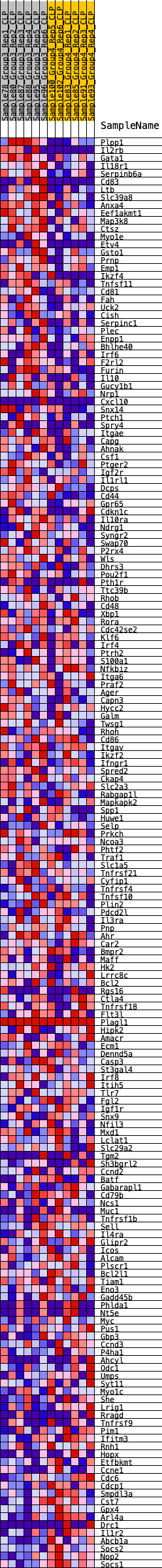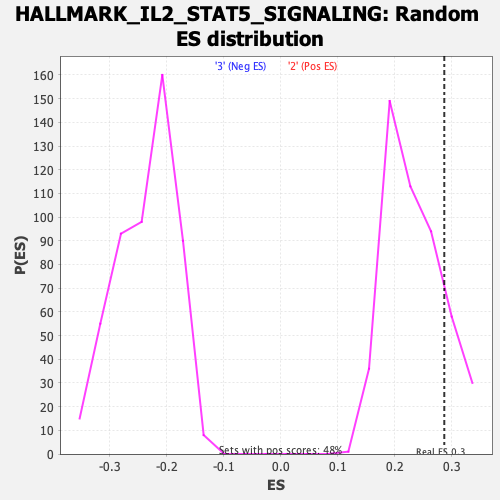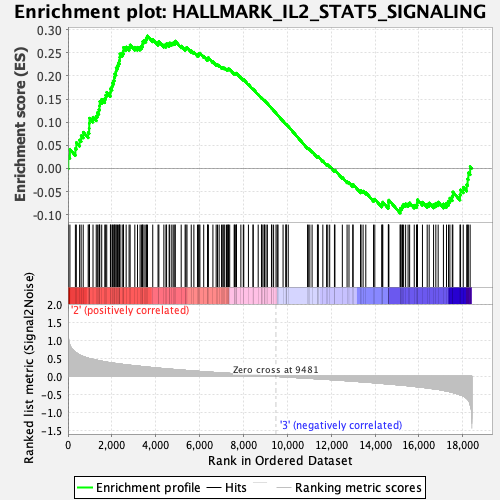
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_IL2_STAT5_SIGNALING |
| Enrichment Score (ES) | 0.28662404 |
| Normalized Enrichment Score (NES) | 1.2315376 |
| Nominal p-value | 0.16839917 |
| FDR q-value | 0.6047192 |
| FWER p-Value | 0.896 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Plpp1 | 14 | 1.147 | 0.0256 | Yes |
| 2 | Il2rb | 87 | 0.862 | 0.0416 | Yes |
| 3 | Gata1 | 337 | 0.668 | 0.0433 | Yes |
| 4 | Il18r1 | 379 | 0.651 | 0.0560 | Yes |
| 5 | Serpinb6a | 529 | 0.590 | 0.0614 | Yes |
| 6 | Cd83 | 599 | 0.572 | 0.0708 | Yes |
| 7 | Ltb | 694 | 0.552 | 0.0784 | Yes |
| 8 | Slc39a8 | 920 | 0.505 | 0.0777 | Yes |
| 9 | Anxa4 | 964 | 0.495 | 0.0867 | Yes |
| 10 | Eef1akmt1 | 970 | 0.494 | 0.0978 | Yes |
| 11 | Map3k8 | 979 | 0.493 | 0.1087 | Yes |
| 12 | Ctsz | 1140 | 0.472 | 0.1108 | Yes |
| 13 | Myo1e | 1287 | 0.449 | 0.1132 | Yes |
| 14 | Etv4 | 1337 | 0.444 | 0.1207 | Yes |
| 15 | Gsto1 | 1408 | 0.437 | 0.1269 | Yes |
| 16 | Prnp | 1437 | 0.432 | 0.1353 | Yes |
| 17 | Emp1 | 1448 | 0.431 | 0.1447 | Yes |
| 18 | Ikzf4 | 1537 | 0.420 | 0.1496 | Yes |
| 19 | Tnfsf11 | 1678 | 0.402 | 0.1511 | Yes |
| 20 | Cd81 | 1717 | 0.398 | 0.1582 | Yes |
| 21 | Fah | 1766 | 0.394 | 0.1647 | Yes |
| 22 | Uck2 | 1934 | 0.377 | 0.1642 | Yes |
| 23 | Cish | 1938 | 0.377 | 0.1727 | Yes |
| 24 | Serpinc1 | 2006 | 0.372 | 0.1776 | Yes |
| 25 | Plec | 2025 | 0.371 | 0.1851 | Yes |
| 26 | Enpp1 | 2086 | 0.366 | 0.1903 | Yes |
| 27 | Bhlhe40 | 2110 | 0.363 | 0.1974 | Yes |
| 28 | Irf6 | 2133 | 0.361 | 0.2045 | Yes |
| 29 | F2rl2 | 2188 | 0.355 | 0.2097 | Yes |
| 30 | Furin | 2195 | 0.355 | 0.2176 | Yes |
| 31 | Il10 | 2255 | 0.348 | 0.2223 | Yes |
| 32 | Gucy1b1 | 2293 | 0.345 | 0.2283 | Yes |
| 33 | Nrp1 | 2342 | 0.340 | 0.2335 | Yes |
| 34 | Cxcl10 | 2359 | 0.339 | 0.2404 | Yes |
| 35 | Snx14 | 2367 | 0.339 | 0.2478 | Yes |
| 36 | Ptch1 | 2471 | 0.334 | 0.2498 | Yes |
| 37 | Spry4 | 2527 | 0.329 | 0.2544 | Yes |
| 38 | Itgae | 2531 | 0.329 | 0.2618 | Yes |
| 39 | Capg | 2651 | 0.318 | 0.2626 | Yes |
| 40 | Ahnak | 2785 | 0.311 | 0.2625 | Yes |
| 41 | Csf1 | 2832 | 0.307 | 0.2670 | Yes |
| 42 | Ptger2 | 3046 | 0.294 | 0.2621 | Yes |
| 43 | Igf2r | 3168 | 0.287 | 0.2621 | Yes |
| 44 | Il1rl1 | 3286 | 0.278 | 0.2621 | Yes |
| 45 | Dcps | 3355 | 0.273 | 0.2647 | Yes |
| 46 | Cd44 | 3400 | 0.270 | 0.2685 | Yes |
| 47 | Gpr65 | 3406 | 0.269 | 0.2744 | Yes |
| 48 | Cdkn1c | 3468 | 0.266 | 0.2772 | Yes |
| 49 | Il10ra | 3551 | 0.261 | 0.2787 | Yes |
| 50 | Ndrg1 | 3582 | 0.258 | 0.2830 | Yes |
| 51 | Syngr2 | 3624 | 0.256 | 0.2866 | Yes |
| 52 | Swap70 | 3863 | 0.244 | 0.2792 | No |
| 53 | P2rx4 | 4105 | 0.229 | 0.2712 | No |
| 54 | Wls | 4142 | 0.227 | 0.2745 | No |
| 55 | Dhrs3 | 4370 | 0.213 | 0.2670 | No |
| 56 | Pou2f1 | 4465 | 0.209 | 0.2666 | No |
| 57 | Pth1r | 4493 | 0.208 | 0.2699 | No |
| 58 | Ttc39b | 4611 | 0.202 | 0.2682 | No |
| 59 | Rhob | 4638 | 0.201 | 0.2714 | No |
| 60 | Cd48 | 4732 | 0.195 | 0.2708 | No |
| 61 | Xbp1 | 4797 | 0.192 | 0.2717 | No |
| 62 | Rora | 4870 | 0.188 | 0.2721 | No |
| 63 | Cdc42se2 | 4888 | 0.187 | 0.2755 | No |
| 64 | Klf6 | 5160 | 0.175 | 0.2646 | No |
| 65 | Irf4 | 5339 | 0.166 | 0.2587 | No |
| 66 | Ptrh2 | 5375 | 0.164 | 0.2605 | No |
| 67 | S100a1 | 5424 | 0.162 | 0.2616 | No |
| 68 | Nfkbiz | 5618 | 0.153 | 0.2546 | No |
| 69 | Itga6 | 5738 | 0.147 | 0.2514 | No |
| 70 | Praf2 | 5902 | 0.141 | 0.2457 | No |
| 71 | Ager | 5959 | 0.139 | 0.2459 | No |
| 72 | Capn3 | 5971 | 0.138 | 0.2484 | No |
| 73 | Hycc2 | 6013 | 0.136 | 0.2493 | No |
| 74 | Galm | 6186 | 0.129 | 0.2428 | No |
| 75 | Twsg1 | 6363 | 0.121 | 0.2360 | No |
| 76 | Rhoh | 6367 | 0.121 | 0.2386 | No |
| 77 | Cd86 | 6400 | 0.120 | 0.2396 | No |
| 78 | Itgav | 6606 | 0.111 | 0.2309 | No |
| 79 | Ikzf2 | 6754 | 0.104 | 0.2252 | No |
| 80 | Ifngr1 | 6814 | 0.101 | 0.2243 | No |
| 81 | Spred2 | 6890 | 0.097 | 0.2224 | No |
| 82 | Ckap4 | 6999 | 0.093 | 0.2187 | No |
| 83 | Slc2a3 | 7055 | 0.091 | 0.2177 | No |
| 84 | Rabgap1l | 7101 | 0.089 | 0.2173 | No |
| 85 | Mapkapk2 | 7145 | 0.087 | 0.2170 | No |
| 86 | Spp1 | 7225 | 0.087 | 0.2146 | No |
| 87 | Huwe1 | 7280 | 0.085 | 0.2136 | No |
| 88 | Selp | 7295 | 0.085 | 0.2148 | No |
| 89 | Prkch | 7315 | 0.084 | 0.2157 | No |
| 90 | Ncoa3 | 7371 | 0.082 | 0.2146 | No |
| 91 | Phtf2 | 7575 | 0.074 | 0.2051 | No |
| 92 | Traf1 | 7595 | 0.073 | 0.2058 | No |
| 93 | Slc1a5 | 7627 | 0.071 | 0.2057 | No |
| 94 | Tnfrsf21 | 7678 | 0.069 | 0.2046 | No |
| 95 | Cyfip1 | 7681 | 0.069 | 0.2061 | No |
| 96 | Tnfrsf4 | 7880 | 0.062 | 0.1966 | No |
| 97 | Tnfsf10 | 7991 | 0.059 | 0.1920 | No |
| 98 | Plin2 | 8013 | 0.057 | 0.1921 | No |
| 99 | Pdcd2l | 8225 | 0.049 | 0.1817 | No |
| 100 | Il3ra | 8434 | 0.039 | 0.1712 | No |
| 101 | Pnp | 8442 | 0.039 | 0.1717 | No |
| 102 | Ahr | 8670 | 0.029 | 0.1599 | No |
| 103 | Car2 | 8819 | 0.024 | 0.1523 | No |
| 104 | Bmpr2 | 8826 | 0.023 | 0.1525 | No |
| 105 | Maff | 8884 | 0.021 | 0.1499 | No |
| 106 | Hk2 | 8948 | 0.019 | 0.1469 | No |
| 107 | Lrrc8c | 8955 | 0.019 | 0.1470 | No |
| 108 | Bcl2 | 9026 | 0.016 | 0.1435 | No |
| 109 | Rgs16 | 9089 | 0.013 | 0.1404 | No |
| 110 | Ctla4 | 9276 | 0.007 | 0.1304 | No |
| 111 | Tnfrsf18 | 9293 | 0.007 | 0.1296 | No |
| 112 | Flt3l | 9372 | 0.004 | 0.1254 | No |
| 113 | Plagl1 | 9483 | 0.000 | 0.1194 | No |
| 114 | Hipk2 | 9539 | -0.000 | 0.1164 | No |
| 115 | Amacr | 9574 | -0.001 | 0.1146 | No |
| 116 | Ecm1 | 9804 | -0.010 | 0.1022 | No |
| 117 | Dennd5a | 9938 | -0.014 | 0.0953 | No |
| 118 | Casp3 | 9943 | -0.015 | 0.0954 | No |
| 119 | St3gal4 | 9944 | -0.015 | 0.0957 | No |
| 120 | Irf8 | 10040 | -0.018 | 0.0909 | No |
| 121 | Itih5 | 10921 | -0.051 | 0.0438 | No |
| 122 | Tlr7 | 10953 | -0.052 | 0.0433 | No |
| 123 | Fgl2 | 11024 | -0.054 | 0.0407 | No |
| 124 | Igf1r | 11127 | -0.057 | 0.0364 | No |
| 125 | Snx9 | 11370 | -0.066 | 0.0247 | No |
| 126 | Nfil3 | 11395 | -0.067 | 0.0249 | No |
| 127 | Mxd1 | 11406 | -0.068 | 0.0259 | No |
| 128 | Lclat1 | 11617 | -0.076 | 0.0161 | No |
| 129 | Slc29a2 | 11788 | -0.082 | 0.0087 | No |
| 130 | Tgm2 | 11842 | -0.084 | 0.0077 | No |
| 131 | Sh3bgrl2 | 11930 | -0.088 | 0.0050 | No |
| 132 | Ccnd2 | 12146 | -0.096 | -0.0046 | No |
| 133 | Batf | 12168 | -0.097 | -0.0035 | No |
| 134 | Gabarapl1 | 12506 | -0.111 | -0.0194 | No |
| 135 | Cd79b | 12722 | -0.120 | -0.0285 | No |
| 136 | Ncs1 | 12809 | -0.123 | -0.0303 | No |
| 137 | Muc1 | 12982 | -0.131 | -0.0368 | No |
| 138 | Tnfrsf1b | 12996 | -0.132 | -0.0344 | No |
| 139 | Sell | 13337 | -0.147 | -0.0497 | No |
| 140 | Il4ra | 13358 | -0.148 | -0.0474 | No |
| 141 | Glipr2 | 13452 | -0.152 | -0.0490 | No |
| 142 | Icos | 13575 | -0.157 | -0.0521 | No |
| 143 | Alcam | 13928 | -0.174 | -0.0674 | No |
| 144 | Plscr1 | 13986 | -0.177 | -0.0664 | No |
| 145 | Bcl2l1 | 14301 | -0.194 | -0.0792 | No |
| 146 | Tiam1 | 14329 | -0.196 | -0.0761 | No |
| 147 | Eno3 | 14350 | -0.196 | -0.0727 | No |
| 148 | Gadd45b | 14602 | -0.207 | -0.0817 | No |
| 149 | Phlda1 | 14611 | -0.208 | -0.0774 | No |
| 150 | Nt5e | 14612 | -0.208 | -0.0726 | No |
| 151 | Myc | 14626 | -0.209 | -0.0685 | No |
| 152 | Pus1 | 15142 | -0.236 | -0.0913 | No |
| 153 | Gbp3 | 15151 | -0.236 | -0.0863 | No |
| 154 | Ccnd3 | 15217 | -0.241 | -0.0843 | No |
| 155 | P4ha1 | 15249 | -0.243 | -0.0804 | No |
| 156 | Ahcyl | 15299 | -0.246 | -0.0774 | No |
| 157 | Odc1 | 15391 | -0.252 | -0.0766 | No |
| 158 | Umps | 15503 | -0.259 | -0.0767 | No |
| 159 | Syt11 | 15574 | -0.264 | -0.0745 | No |
| 160 | Myo1c | 15784 | -0.278 | -0.0795 | No |
| 161 | She | 15886 | -0.286 | -0.0785 | No |
| 162 | Lrig1 | 15922 | -0.288 | -0.0738 | No |
| 163 | Rragd | 15932 | -0.289 | -0.0676 | No |
| 164 | Tnfrsf9 | 16158 | -0.305 | -0.0729 | No |
| 165 | Pim1 | 16374 | -0.320 | -0.0773 | No |
| 166 | Ifitm3 | 16464 | -0.328 | -0.0747 | No |
| 167 | Rnh1 | 16665 | -0.344 | -0.0777 | No |
| 168 | Hopx | 16768 | -0.352 | -0.0752 | No |
| 169 | Etfbkmt | 16874 | -0.361 | -0.0727 | No |
| 170 | Ccne1 | 17114 | -0.389 | -0.0768 | No |
| 171 | Cdc6 | 17254 | -0.407 | -0.0751 | No |
| 172 | Cdcp1 | 17348 | -0.418 | -0.0706 | No |
| 173 | Smpdl3a | 17409 | -0.424 | -0.0641 | No |
| 174 | Cst7 | 17518 | -0.443 | -0.0598 | No |
| 175 | Gpx4 | 17543 | -0.447 | -0.0508 | No |
| 176 | Arl4a | 17869 | -0.509 | -0.0569 | No |
| 177 | Drc1 | 17895 | -0.516 | -0.0464 | No |
| 178 | Il1r2 | 18017 | -0.542 | -0.0406 | No |
| 179 | Abcb1a | 18177 | -0.609 | -0.0353 | No |
| 180 | Socs2 | 18217 | -0.633 | -0.0228 | No |
| 181 | Nop2 | 18250 | -0.656 | -0.0095 | No |
| 182 | Socs1 | 18332 | -0.785 | 0.0042 | No |