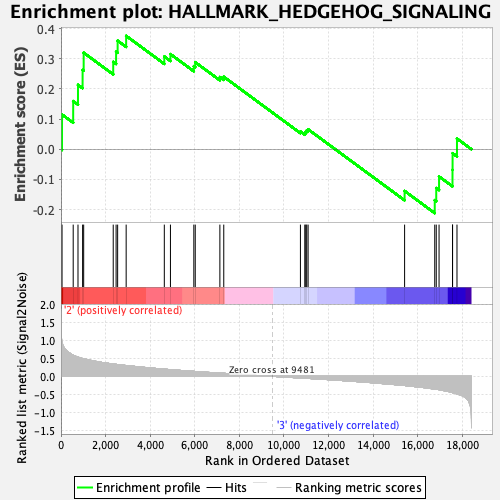
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.37602714 |
| Normalized Enrichment Score (NES) | 1.2923152 |
| Nominal p-value | 0.09330629 |
| FDR q-value | 0.58293456 |
| FWER p-Value | 0.817 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
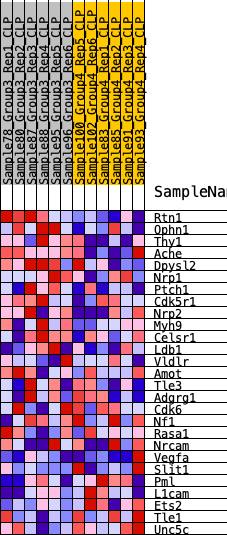
| 1 | Rtn1 | 46 | 0.962 | 0.1152 | Yes |
| 2 | Ophn1 | 547 | 0.586 | 0.1597 | Yes |
| 3 | Thy1 | 762 | 0.536 | 0.2137 | Yes |
| 4 | Ache | 969 | 0.494 | 0.2629 | Yes |
| 5 | Dpysl2 | 1017 | 0.489 | 0.3202 | Yes |
| 6 | Nrp1 | 2342 | 0.340 | 0.2897 | Yes |
| 7 | Ptch1 | 2471 | 0.334 | 0.3236 | Yes |
| 8 | Cdk5r1 | 2540 | 0.328 | 0.3600 | Yes |
| 9 | Nrp2 | 2922 | 0.300 | 0.3760 | Yes |
| 10 | Myh9 | 4633 | 0.201 | 0.3076 | No |
| 11 | Celsr1 | 4908 | 0.186 | 0.3155 | No |
| 12 | Ldb1 | 5957 | 0.139 | 0.2755 | No |
| 13 | Vldlr | 6020 | 0.136 | 0.2887 | No |
| 14 | Amot | 7125 | 0.088 | 0.2395 | No |
| 15 | Tle3 | 7299 | 0.085 | 0.2404 | No |
| 16 | Adgrg1 | 10737 | -0.044 | 0.0588 | No |
| 17 | Cdk6 | 10933 | -0.051 | 0.0544 | No |
| 18 | Nf1 | 10976 | -0.053 | 0.0586 | No |
| 19 | Rasa1 | 11019 | -0.053 | 0.0628 | No |
| 20 | Nrcam | 11081 | -0.055 | 0.0662 | No |
| 21 | Vegfa | 15409 | -0.253 | -0.1382 | No |
| 22 | Slit1 | 16761 | -0.352 | -0.1687 | No |
| 23 | Pml | 16828 | -0.358 | -0.1285 | No |
| 24 | L1cam | 16954 | -0.368 | -0.0903 | No |
| 25 | Ets2 | 17557 | -0.449 | -0.0681 | No |
| 26 | Tle1 | 17561 | -0.451 | -0.0131 | No |
| 27 | Unc5c | 17758 | -0.484 | 0.0354 | No |