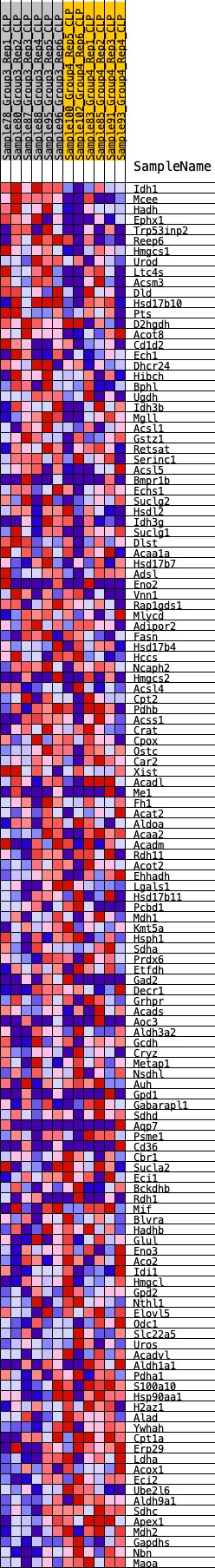
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
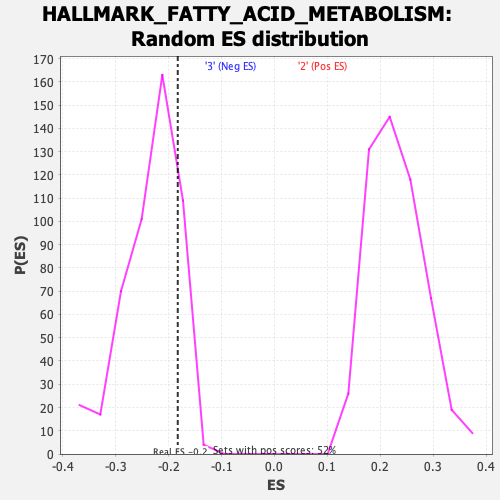
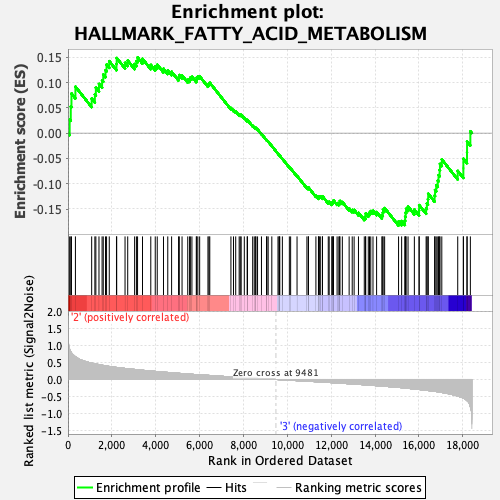
| Enrichment Score (ES) | -0.18279202 |
| Normalized Enrichment Score (NES) | -0.7897597 |
| Nominal p-value | 0.82061857 |
| FDR q-value | 0.957957 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idh1 | 75 | 0.885 | 0.0269 | No |
| 2 | Mcee | 126 | 0.811 | 0.0526 | No |
| 3 | Hadh | 158 | 0.779 | 0.0783 | No |
| 4 | Ephx1 | 338 | 0.666 | 0.0918 | No |
| 5 | Trp53inp2 | 1079 | 0.482 | 0.0682 | No |
| 6 | Reep6 | 1225 | 0.457 | 0.0763 | No |
| 7 | Hmgcs1 | 1270 | 0.451 | 0.0897 | No |
| 8 | Urod | 1412 | 0.436 | 0.0973 | No |
| 9 | Ltc4s | 1554 | 0.418 | 0.1042 | No |
| 10 | Acsm3 | 1607 | 0.412 | 0.1158 | No |
| 11 | Dld | 1709 | 0.398 | 0.1243 | No |
| 12 | Hsd17b10 | 1758 | 0.395 | 0.1355 | No |
| 13 | Pts | 1881 | 0.382 | 0.1422 | No |
| 14 | D2hgdh | 2210 | 0.352 | 0.1366 | No |
| 15 | Acot8 | 2220 | 0.352 | 0.1485 | No |
| 16 | Cd1d2 | 2600 | 0.323 | 0.1390 | No |
| 17 | Ech1 | 2722 | 0.314 | 0.1434 | No |
| 18 | Dhcr24 | 3037 | 0.295 | 0.1366 | No |
| 19 | Hibch | 3119 | 0.290 | 0.1423 | No |
| 20 | Bphl | 3171 | 0.287 | 0.1496 | No |
| 21 | Ugdh | 3396 | 0.270 | 0.1468 | No |
| 22 | Idh3b | 3774 | 0.248 | 0.1349 | No |
| 23 | Mgll | 3977 | 0.236 | 0.1321 | No |
| 24 | Acsl1 | 4068 | 0.231 | 0.1353 | No |
| 25 | Gstz1 | 4353 | 0.214 | 0.1273 | No |
| 26 | Retsat | 4548 | 0.206 | 0.1239 | No |
| 27 | Serinc1 | 4723 | 0.196 | 0.1212 | No |
| 28 | Acsl5 | 5040 | 0.181 | 0.1103 | No |
| 29 | Bmpr1b | 5073 | 0.179 | 0.1148 | No |
| 30 | Echs1 | 5199 | 0.173 | 0.1141 | No |
| 31 | Suclg2 | 5459 | 0.160 | 0.1055 | No |
| 32 | Hsdl2 | 5542 | 0.156 | 0.1065 | No |
| 33 | Idh3g | 5578 | 0.154 | 0.1100 | No |
| 34 | Suclg1 | 5649 | 0.151 | 0.1115 | No |
| 35 | Dlst | 5848 | 0.144 | 0.1057 | No |
| 36 | Acaa1a | 5866 | 0.143 | 0.1098 | No |
| 37 | Hsd17b7 | 5919 | 0.140 | 0.1118 | No |
| 38 | Adsl | 6002 | 0.136 | 0.1121 | No |
| 39 | Eno2 | 6380 | 0.120 | 0.0957 | No |
| 40 | Vnn1 | 6425 | 0.119 | 0.0975 | No |
| 41 | Rap1gds1 | 6463 | 0.117 | 0.0995 | No |
| 42 | Mlycd | 7427 | 0.079 | 0.0496 | No |
| 43 | Adipor2 | 7544 | 0.075 | 0.0459 | No |
| 44 | Fasn | 7645 | 0.071 | 0.0429 | No |
| 45 | Hsd17b4 | 7803 | 0.065 | 0.0366 | No |
| 46 | Hccs | 7852 | 0.063 | 0.0362 | No |
| 47 | Ncaph2 | 7894 | 0.061 | 0.0361 | No |
| 48 | Hmgcs2 | 8037 | 0.057 | 0.0303 | No |
| 49 | Acsl4 | 8168 | 0.051 | 0.0250 | No |
| 50 | Cpt2 | 8181 | 0.051 | 0.0261 | No |
| 51 | Pdhb | 8419 | 0.040 | 0.0145 | No |
| 52 | Acss1 | 8491 | 0.037 | 0.0119 | No |
| 53 | Crat | 8555 | 0.034 | 0.0097 | No |
| 54 | Cpox | 8565 | 0.034 | 0.0103 | No |
| 55 | Ostc | 8641 | 0.030 | 0.0073 | No |
| 56 | Car2 | 8819 | 0.024 | -0.0015 | No |
| 57 | Xist | 9050 | 0.015 | -0.0136 | No |
| 58 | Acadl | 9067 | 0.015 | -0.0140 | No |
| 59 | Me1 | 9124 | 0.012 | -0.0166 | No |
| 60 | Fh1 | 9291 | 0.007 | -0.0255 | No |
| 61 | Acat2 | 9573 | -0.001 | -0.0408 | No |
| 62 | Aldoa | 9618 | -0.003 | -0.0431 | No |
| 63 | Acaa2 | 9639 | -0.004 | -0.0441 | No |
| 64 | Acadm | 9655 | -0.004 | -0.0447 | No |
| 65 | Rdh11 | 9772 | -0.009 | -0.0508 | No |
| 66 | Acot2 | 10090 | -0.020 | -0.0674 | No |
| 67 | Ehhadh | 10149 | -0.022 | -0.0698 | No |
| 68 | Lgals1 | 10441 | -0.033 | -0.0846 | No |
| 69 | Hsd17b11 | 10891 | -0.050 | -0.1074 | No |
| 70 | Pcbd1 | 10956 | -0.052 | -0.1091 | No |
| 71 | Mdh1 | 10961 | -0.052 | -0.1075 | No |
| 72 | Kmt5a | 11301 | -0.064 | -0.1238 | No |
| 73 | Hsph1 | 11417 | -0.068 | -0.1277 | No |
| 74 | Sdha | 11422 | -0.068 | -0.1256 | No |
| 75 | Prdx6 | 11459 | -0.069 | -0.1251 | No |
| 76 | Etfdh | 11501 | -0.071 | -0.1248 | No |
| 77 | Gad2 | 11596 | -0.075 | -0.1274 | No |
| 78 | Decr1 | 11600 | -0.075 | -0.1249 | No |
| 79 | Grhpr | 11864 | -0.085 | -0.1363 | No |
| 80 | Acads | 11906 | -0.086 | -0.1355 | No |
| 81 | Aoc3 | 12025 | -0.092 | -0.1387 | No |
| 82 | Aldh3a2 | 12049 | -0.093 | -0.1367 | No |
| 83 | Gcdh | 12083 | -0.094 | -0.1353 | No |
| 84 | Cryz | 12103 | -0.095 | -0.1330 | No |
| 85 | Metap1 | 12267 | -0.101 | -0.1383 | No |
| 86 | Nsdhl | 12347 | -0.104 | -0.1390 | No |
| 87 | Auh | 12384 | -0.107 | -0.1372 | No |
| 88 | Gpd1 | 12394 | -0.107 | -0.1340 | No |
| 89 | Gabarapl1 | 12506 | -0.111 | -0.1362 | No |
| 90 | Sdhd | 12816 | -0.124 | -0.1487 | No |
| 91 | Aqp7 | 12956 | -0.130 | -0.1518 | No |
| 92 | Psme1 | 13043 | -0.134 | -0.1518 | No |
| 93 | Cd36 | 13241 | -0.143 | -0.1575 | No |
| 94 | Cbr1 | 13506 | -0.155 | -0.1665 | No |
| 95 | Sucla2 | 13566 | -0.157 | -0.1643 | No |
| 96 | Eci1 | 13569 | -0.157 | -0.1589 | No |
| 97 | Bckdhb | 13689 | -0.164 | -0.1596 | No |
| 98 | Rdh1 | 13744 | -0.166 | -0.1568 | No |
| 99 | Mif | 13802 | -0.169 | -0.1540 | No |
| 100 | Blvra | 13903 | -0.173 | -0.1534 | No |
| 101 | Hadhb | 14068 | -0.182 | -0.1560 | No |
| 102 | Glul | 14307 | -0.194 | -0.1622 | No |
| 103 | Eno3 | 14350 | -0.196 | -0.1576 | No |
| 104 | Aco2 | 14360 | -0.197 | -0.1511 | No |
| 105 | Idi1 | 14434 | -0.201 | -0.1481 | No |
| 106 | Hmgcl | 15069 | -0.234 | -0.1746 | Yes |
| 107 | Gpd2 | 15209 | -0.240 | -0.1737 | Yes |
| 108 | Nthl1 | 15357 | -0.250 | -0.1730 | Yes |
| 109 | Elovl5 | 15371 | -0.251 | -0.1649 | Yes |
| 110 | Odc1 | 15391 | -0.252 | -0.1571 | Yes |
| 111 | Slc22a5 | 15413 | -0.253 | -0.1494 | Yes |
| 112 | Uros | 15501 | -0.259 | -0.1450 | Yes |
| 113 | Acadvl | 15789 | -0.279 | -0.1510 | Yes |
| 114 | Aldh1a1 | 16004 | -0.293 | -0.1524 | Yes |
| 115 | Pdha1 | 16008 | -0.293 | -0.1423 | Yes |
| 116 | S100a10 | 16325 | -0.315 | -0.1485 | Yes |
| 117 | Hsp90aa1 | 16359 | -0.318 | -0.1392 | Yes |
| 118 | H2az1 | 16409 | -0.323 | -0.1305 | Yes |
| 119 | Alad | 16420 | -0.324 | -0.1197 | Yes |
| 120 | Ywhah | 16711 | -0.347 | -0.1234 | Yes |
| 121 | Cpt1a | 16733 | -0.349 | -0.1123 | Yes |
| 122 | Erp29 | 16786 | -0.353 | -0.1028 | Yes |
| 123 | Ldha | 16857 | -0.359 | -0.0940 | Yes |
| 124 | Acox1 | 16891 | -0.362 | -0.0831 | Yes |
| 125 | Eci2 | 16941 | -0.367 | -0.0729 | Yes |
| 126 | Ube2l6 | 16956 | -0.368 | -0.0608 | Yes |
| 127 | Aldh9a1 | 17040 | -0.379 | -0.0520 | Yes |
| 128 | Sdhc | 17766 | -0.485 | -0.0747 | Yes |
| 129 | Apex1 | 18023 | -0.543 | -0.0696 | Yes |
| 130 | Mdh2 | 18024 | -0.544 | -0.0506 | Yes |
| 131 | Gapdhs | 18189 | -0.617 | -0.0379 | Yes |
| 132 | Nbn | 18192 | -0.618 | -0.0163 | Yes |
| 133 | Maoa | 18342 | -0.801 | 0.0036 | Yes |