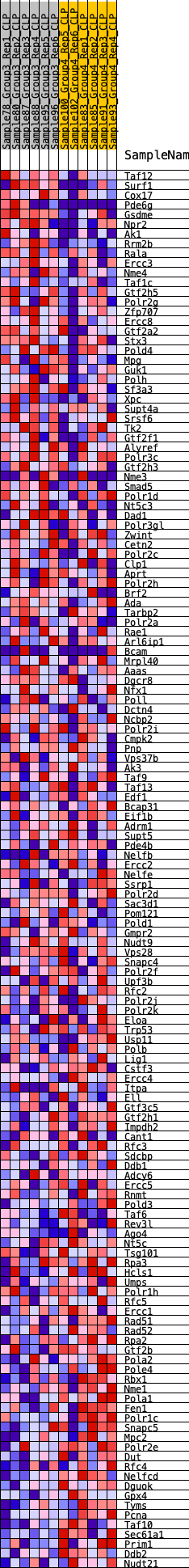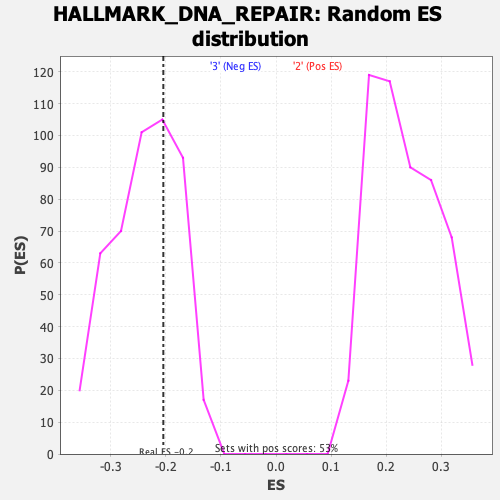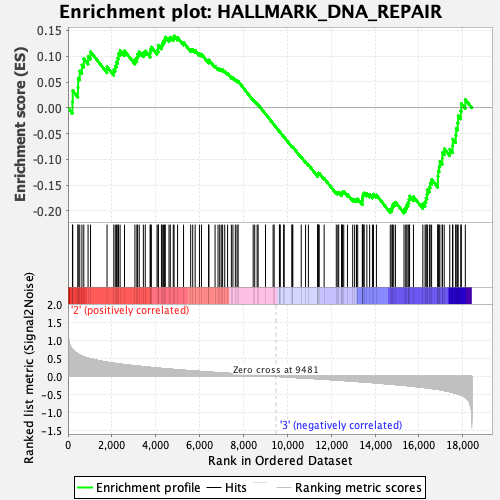
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_DNA_REPAIR |
| Enrichment Score (ES) | -0.20425265 |
| Normalized Enrichment Score (NES) | -0.86072993 |
| Nominal p-value | 0.68443495 |
| FDR q-value | 0.99291265 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Taf12 | 203 | 0.743 | 0.0115 | No |
| 2 | Surf1 | 217 | 0.735 | 0.0332 | No |
| 3 | Cox17 | 453 | 0.618 | 0.0391 | No |
| 4 | Pde6g | 461 | 0.615 | 0.0575 | No |
| 5 | Gsdme | 535 | 0.589 | 0.0714 | No |
| 6 | Npr2 | 635 | 0.563 | 0.0831 | No |
| 7 | Ak1 | 720 | 0.546 | 0.0952 | No |
| 8 | Rrm2b | 915 | 0.507 | 0.1000 | No |
| 9 | Rala | 1027 | 0.488 | 0.1088 | No |
| 10 | Ercc3 | 1779 | 0.393 | 0.0796 | No |
| 11 | Nme4 | 2088 | 0.366 | 0.0739 | No |
| 12 | Taf1c | 2155 | 0.359 | 0.0813 | No |
| 13 | Gtf2h5 | 2208 | 0.352 | 0.0891 | No |
| 14 | Polr2g | 2269 | 0.346 | 0.0964 | No |
| 15 | Zfp707 | 2302 | 0.344 | 0.1051 | No |
| 16 | Ercc8 | 2375 | 0.338 | 0.1115 | No |
| 17 | Gtf2a2 | 2575 | 0.325 | 0.1105 | No |
| 18 | Stx3 | 3051 | 0.294 | 0.0935 | No |
| 19 | Pold4 | 3139 | 0.289 | 0.0975 | No |
| 20 | Mpg | 3174 | 0.287 | 0.1044 | No |
| 21 | Guk1 | 3247 | 0.281 | 0.1090 | No |
| 22 | Polh | 3435 | 0.268 | 0.1069 | No |
| 23 | Sf3a3 | 3522 | 0.264 | 0.1103 | No |
| 24 | Xpc | 3748 | 0.249 | 0.1055 | No |
| 25 | Supt4a | 3752 | 0.249 | 0.1129 | No |
| 26 | Srsf6 | 3800 | 0.246 | 0.1179 | No |
| 27 | Tk2 | 4059 | 0.232 | 0.1108 | No |
| 28 | Gtf2f1 | 4113 | 0.228 | 0.1149 | No |
| 29 | Alyref | 4118 | 0.228 | 0.1216 | No |
| 30 | Polr3c | 4259 | 0.220 | 0.1206 | No |
| 31 | Gtf2h3 | 4295 | 0.217 | 0.1253 | No |
| 32 | Nme3 | 4349 | 0.214 | 0.1289 | No |
| 33 | Smad5 | 4404 | 0.213 | 0.1324 | No |
| 34 | Polr1d | 4434 | 0.211 | 0.1373 | No |
| 35 | Nt5c3 | 4605 | 0.203 | 0.1342 | No |
| 36 | Dad1 | 4663 | 0.200 | 0.1371 | No |
| 37 | Polr3gl | 4798 | 0.192 | 0.1356 | No |
| 38 | Zwint | 4835 | 0.190 | 0.1395 | No |
| 39 | Cetn2 | 4995 | 0.183 | 0.1363 | No |
| 40 | Polr2c | 5267 | 0.170 | 0.1267 | No |
| 41 | Clp1 | 5593 | 0.153 | 0.1135 | No |
| 42 | Aprt | 5685 | 0.149 | 0.1131 | No |
| 43 | Polr2h | 5796 | 0.145 | 0.1115 | No |
| 44 | Brf2 | 5992 | 0.137 | 0.1050 | No |
| 45 | Ada | 6084 | 0.133 | 0.1041 | No |
| 46 | Tarbp2 | 6408 | 0.119 | 0.0900 | No |
| 47 | Polr2a | 6423 | 0.119 | 0.0928 | No |
| 48 | Rae1 | 6704 | 0.107 | 0.0808 | No |
| 49 | Arl6ip1 | 6839 | 0.100 | 0.0765 | No |
| 50 | Bcam | 6916 | 0.097 | 0.0753 | No |
| 51 | Mrpl40 | 6986 | 0.094 | 0.0743 | No |
| 52 | Aaas | 7049 | 0.091 | 0.0737 | No |
| 53 | Dgcr8 | 7144 | 0.087 | 0.0712 | No |
| 54 | Nfx1 | 7275 | 0.086 | 0.0667 | No |
| 55 | Poll | 7447 | 0.079 | 0.0597 | No |
| 56 | Dctn4 | 7518 | 0.076 | 0.0582 | No |
| 57 | Ncbp2 | 7628 | 0.071 | 0.0544 | No |
| 58 | Polr2i | 7705 | 0.068 | 0.0523 | No |
| 59 | Cmpk2 | 7756 | 0.066 | 0.0516 | No |
| 60 | Pnp | 8442 | 0.039 | 0.0153 | No |
| 61 | Vps37b | 8512 | 0.036 | 0.0126 | No |
| 62 | Ak3 | 8622 | 0.031 | 0.0076 | No |
| 63 | Taf9 | 8666 | 0.029 | 0.0061 | No |
| 64 | Taf13 | 9003 | 0.017 | -0.0118 | No |
| 65 | Edf1 | 9350 | 0.004 | -0.0306 | No |
| 66 | Bcap31 | 9398 | 0.003 | -0.0331 | No |
| 67 | Eif1b | 9638 | -0.004 | -0.0461 | No |
| 68 | Adrm1 | 9649 | -0.004 | -0.0465 | No |
| 69 | Supt5 | 9676 | -0.005 | -0.0477 | No |
| 70 | Pde4b | 9829 | -0.011 | -0.0557 | No |
| 71 | Nelfb | 9855 | -0.012 | -0.0567 | No |
| 72 | Ercc2 | 10203 | -0.024 | -0.0750 | No |
| 73 | Nelfe | 10227 | -0.024 | -0.0755 | No |
| 74 | Ssrp1 | 10249 | -0.026 | -0.0759 | No |
| 75 | Polr2d | 10632 | -0.040 | -0.0956 | No |
| 76 | Sac3d1 | 10829 | -0.047 | -0.1049 | No |
| 77 | Pom121 | 10958 | -0.052 | -0.1103 | No |
| 78 | Pold1 | 11389 | -0.067 | -0.1318 | No |
| 79 | Gmpr2 | 11392 | -0.067 | -0.1299 | No |
| 80 | Nudt9 | 11397 | -0.067 | -0.1281 | No |
| 81 | Vps28 | 11404 | -0.067 | -0.1263 | No |
| 82 | Snapc4 | 11452 | -0.069 | -0.1268 | No |
| 83 | Polr2f | 11676 | -0.078 | -0.1366 | No |
| 84 | Upf3b | 12230 | -0.100 | -0.1639 | No |
| 85 | Rfc2 | 12300 | -0.103 | -0.1645 | No |
| 86 | Polr2j | 12340 | -0.104 | -0.1635 | No |
| 87 | Polr2k | 12473 | -0.110 | -0.1674 | No |
| 88 | Eloa | 12475 | -0.110 | -0.1641 | No |
| 89 | Trp53 | 12516 | -0.112 | -0.1629 | No |
| 90 | Usp11 | 12570 | -0.114 | -0.1623 | No |
| 91 | Polb | 12739 | -0.121 | -0.1678 | No |
| 92 | Lig1 | 12978 | -0.131 | -0.1769 | No |
| 93 | Cstf3 | 13066 | -0.135 | -0.1775 | No |
| 94 | Ercc4 | 13151 | -0.139 | -0.1779 | No |
| 95 | Itpa | 13206 | -0.142 | -0.1765 | No |
| 96 | Ell | 13419 | -0.150 | -0.1836 | No |
| 97 | Gtf3c5 | 13421 | -0.150 | -0.1790 | No |
| 98 | Gtf2h1 | 13430 | -0.151 | -0.1749 | No |
| 99 | Impdh2 | 13441 | -0.151 | -0.1708 | No |
| 100 | Cant1 | 13453 | -0.152 | -0.1668 | No |
| 101 | Rfc3 | 13504 | -0.155 | -0.1649 | No |
| 102 | Sdcbp | 13624 | -0.160 | -0.1665 | No |
| 103 | Ddb1 | 13750 | -0.166 | -0.1683 | No |
| 104 | Adcy6 | 13877 | -0.172 | -0.1699 | No |
| 105 | Ercc5 | 13925 | -0.174 | -0.1672 | No |
| 106 | Rnmt | 14059 | -0.181 | -0.1689 | No |
| 107 | Pold3 | 14691 | -0.212 | -0.1970 | Yes |
| 108 | Taf6 | 14764 | -0.216 | -0.1944 | Yes |
| 109 | Rev3l | 14788 | -0.217 | -0.1890 | Yes |
| 110 | Ago4 | 14848 | -0.221 | -0.1855 | Yes |
| 111 | Nt5c | 14926 | -0.226 | -0.1828 | Yes |
| 112 | Tsg101 | 15318 | -0.247 | -0.1967 | Yes |
| 113 | Rpa3 | 15396 | -0.253 | -0.1932 | Yes |
| 114 | Hcls1 | 15446 | -0.254 | -0.1882 | Yes |
| 115 | Umps | 15503 | -0.259 | -0.1833 | Yes |
| 116 | Polr1h | 15541 | -0.262 | -0.1774 | Yes |
| 117 | Rfc5 | 15571 | -0.264 | -0.1709 | Yes |
| 118 | Ercc1 | 15747 | -0.275 | -0.1721 | Yes |
| 119 | Rad51 | 16176 | -0.306 | -0.1862 | Yes |
| 120 | Rad52 | 16278 | -0.312 | -0.1823 | Yes |
| 121 | Rpa2 | 16320 | -0.315 | -0.1749 | Yes |
| 122 | Gtf2b | 16365 | -0.319 | -0.1676 | Yes |
| 123 | Pola2 | 16379 | -0.320 | -0.1586 | Yes |
| 124 | Pole4 | 16474 | -0.329 | -0.1537 | Yes |
| 125 | Rbx1 | 16509 | -0.331 | -0.1455 | Yes |
| 126 | Nme1 | 16575 | -0.337 | -0.1388 | Yes |
| 127 | Pola1 | 16860 | -0.359 | -0.1434 | Yes |
| 128 | Fen1 | 16861 | -0.359 | -0.1325 | Yes |
| 129 | Polr1c | 16878 | -0.361 | -0.1223 | Yes |
| 130 | Snapc5 | 16919 | -0.365 | -0.1134 | Yes |
| 131 | Mpc2 | 16952 | -0.368 | -0.1039 | Yes |
| 132 | Polr2e | 17060 | -0.382 | -0.0982 | Yes |
| 133 | Dut | 17065 | -0.382 | -0.0868 | Yes |
| 134 | Rfc4 | 17150 | -0.393 | -0.0794 | Yes |
| 135 | Nelfcd | 17402 | -0.424 | -0.0802 | Yes |
| 136 | Dguok | 17531 | -0.445 | -0.0737 | Yes |
| 137 | Gpx4 | 17543 | -0.447 | -0.0606 | Yes |
| 138 | Tyms | 17672 | -0.471 | -0.0533 | Yes |
| 139 | Pcna | 17694 | -0.473 | -0.0401 | Yes |
| 140 | Taf10 | 17762 | -0.484 | -0.0290 | Yes |
| 141 | Sec61a1 | 17784 | -0.489 | -0.0152 | Yes |
| 142 | Prim1 | 17905 | -0.517 | -0.0061 | Yes |
| 143 | Ddb2 | 17929 | -0.520 | 0.0085 | Yes |
| 144 | Nudt21 | 18112 | -0.579 | 0.0162 | Yes |