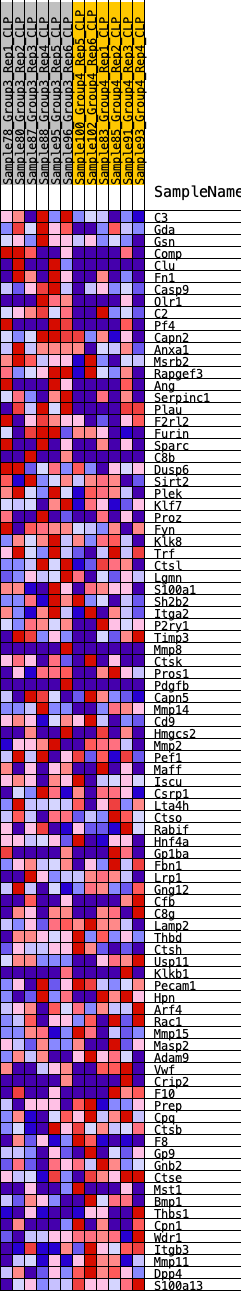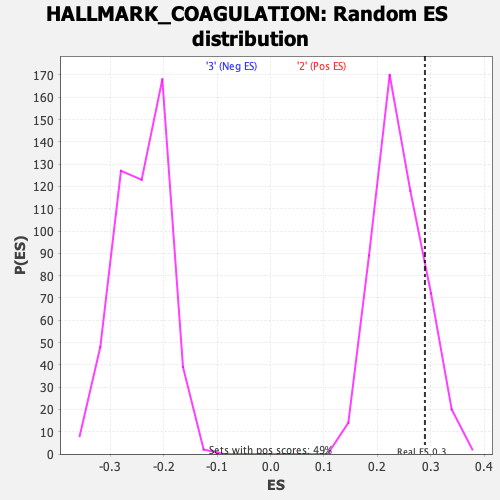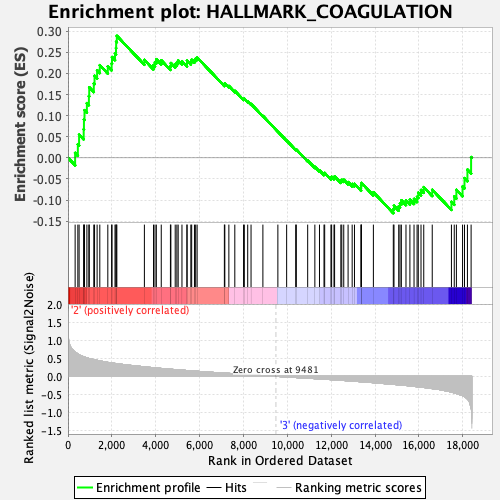
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.2891096 |
| Normalized Enrichment Score (NES) | 1.2052146 |
| Nominal p-value | 0.15670103 |
| FDR q-value | 0.6058962 |
| FWER p-Value | 0.918 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | C3 | 325 | 0.674 | 0.0117 | Yes |
| 2 | Gda | 449 | 0.622 | 0.0321 | Yes |
| 3 | Gsn | 506 | 0.597 | 0.0550 | Yes |
| 4 | Comp | 715 | 0.547 | 0.0675 | Yes |
| 5 | Clu | 727 | 0.545 | 0.0907 | Yes |
| 6 | Fn1 | 752 | 0.539 | 0.1129 | Yes |
| 7 | Casp9 | 861 | 0.518 | 0.1296 | Yes |
| 8 | Olr1 | 953 | 0.498 | 0.1464 | Yes |
| 9 | C2 | 968 | 0.494 | 0.1672 | Yes |
| 10 | Pf4 | 1176 | 0.466 | 0.1762 | Yes |
| 11 | Capn2 | 1210 | 0.459 | 0.1944 | Yes |
| 12 | Anxa1 | 1327 | 0.445 | 0.2075 | Yes |
| 13 | Msrb2 | 1450 | 0.430 | 0.2196 | Yes |
| 14 | Rapgef3 | 1815 | 0.390 | 0.2167 | Yes |
| 15 | Ang | 1989 | 0.373 | 0.2235 | Yes |
| 16 | Serpinc1 | 2006 | 0.372 | 0.2389 | Yes |
| 17 | Plau | 2143 | 0.360 | 0.2472 | Yes |
| 18 | F2rl2 | 2188 | 0.355 | 0.2603 | Yes |
| 19 | Furin | 2195 | 0.355 | 0.2754 | Yes |
| 20 | Sparc | 2226 | 0.352 | 0.2891 | Yes |
| 21 | C8b | 3482 | 0.265 | 0.2321 | No |
| 22 | Dusp6 | 3898 | 0.242 | 0.2201 | No |
| 23 | Sirt2 | 3965 | 0.238 | 0.2268 | No |
| 24 | Plek | 4027 | 0.234 | 0.2337 | No |
| 25 | Klf7 | 4251 | 0.220 | 0.2311 | No |
| 26 | Proz | 4669 | 0.199 | 0.2170 | No |
| 27 | Fyn | 4689 | 0.198 | 0.2246 | No |
| 28 | Klk8 | 4887 | 0.187 | 0.2220 | No |
| 29 | Trf | 4963 | 0.184 | 0.2260 | No |
| 30 | Ctsl | 5022 | 0.182 | 0.2308 | No |
| 31 | Lgmn | 5195 | 0.173 | 0.2289 | No |
| 32 | S100a1 | 5424 | 0.162 | 0.2235 | No |
| 33 | Sh2b2 | 5425 | 0.162 | 0.2306 | No |
| 34 | Itga2 | 5600 | 0.153 | 0.2278 | No |
| 35 | P2ry1 | 5638 | 0.152 | 0.2324 | No |
| 36 | Timp3 | 5765 | 0.146 | 0.2319 | No |
| 37 | Mmp8 | 5820 | 0.144 | 0.2352 | No |
| 38 | Ctsk | 5886 | 0.142 | 0.2379 | No |
| 39 | Pros1 | 7128 | 0.088 | 0.1740 | No |
| 40 | Pdgfb | 7149 | 0.087 | 0.1767 | No |
| 41 | Capn5 | 7333 | 0.083 | 0.1703 | No |
| 42 | Mmp14 | 7604 | 0.073 | 0.1588 | No |
| 43 | Cd9 | 8003 | 0.058 | 0.1396 | No |
| 44 | Hmgcs2 | 8037 | 0.057 | 0.1402 | No |
| 45 | Mmp2 | 8192 | 0.050 | 0.1340 | No |
| 46 | Pef1 | 8344 | 0.044 | 0.1277 | No |
| 47 | Maff | 8884 | 0.021 | 0.0992 | No |
| 48 | Iscu | 9567 | -0.001 | 0.0620 | No |
| 49 | Csrp1 | 9967 | -0.016 | 0.0409 | No |
| 50 | Lta4h | 10380 | -0.031 | 0.0198 | No |
| 51 | Ctso | 10408 | -0.032 | 0.0197 | No |
| 52 | Rabif | 10926 | -0.051 | -0.0063 | No |
| 53 | Hnf4a | 11253 | -0.062 | -0.0214 | No |
| 54 | Gp1ba | 11464 | -0.070 | -0.0298 | No |
| 55 | Fbn1 | 11670 | -0.077 | -0.0376 | No |
| 56 | Lrp1 | 11701 | -0.079 | -0.0358 | No |
| 57 | Gng12 | 11987 | -0.090 | -0.0474 | No |
| 58 | Cfb | 12018 | -0.092 | -0.0451 | No |
| 59 | C8g | 12126 | -0.096 | -0.0468 | No |
| 60 | Lamp2 | 12153 | -0.097 | -0.0440 | No |
| 61 | Thbd | 12438 | -0.108 | -0.0547 | No |
| 62 | Ctsh | 12481 | -0.110 | -0.0522 | No |
| 63 | Usp11 | 12570 | -0.114 | -0.0521 | No |
| 64 | Klkb1 | 12768 | -0.122 | -0.0575 | No |
| 65 | Pecam1 | 12954 | -0.130 | -0.0619 | No |
| 66 | Hpn | 13059 | -0.135 | -0.0617 | No |
| 67 | Arf4 | 13360 | -0.148 | -0.0717 | No |
| 68 | Rac1 | 13362 | -0.148 | -0.0653 | No |
| 69 | Mmp15 | 13377 | -0.148 | -0.0595 | No |
| 70 | Masp2 | 13921 | -0.174 | -0.0816 | No |
| 71 | Adam9 | 14829 | -0.220 | -0.1215 | No |
| 72 | Vwf | 14860 | -0.222 | -0.1135 | No |
| 73 | Crip2 | 15076 | -0.235 | -0.1150 | No |
| 74 | F10 | 15134 | -0.235 | -0.1078 | No |
| 75 | Prep | 15196 | -0.239 | -0.1007 | No |
| 76 | Cpq | 15407 | -0.253 | -0.1011 | No |
| 77 | Ctsb | 15586 | -0.265 | -0.0993 | No |
| 78 | F8 | 15774 | -0.277 | -0.0974 | No |
| 79 | Gp9 | 15913 | -0.288 | -0.0924 | No |
| 80 | Gnb2 | 15968 | -0.292 | -0.0826 | No |
| 81 | Ctse | 16092 | -0.300 | -0.0762 | No |
| 82 | Mst1 | 16217 | -0.309 | -0.0695 | No |
| 83 | Bmp1 | 16604 | -0.340 | -0.0757 | No |
| 84 | Thbs1 | 17479 | -0.436 | -0.1044 | No |
| 85 | Cpn1 | 17608 | -0.459 | -0.0914 | No |
| 86 | Wdr1 | 17705 | -0.475 | -0.0759 | No |
| 87 | Itgb3 | 17986 | -0.534 | -0.0679 | No |
| 88 | Mmp11 | 18074 | -0.562 | -0.0481 | No |
| 89 | Dpp4 | 18208 | -0.629 | -0.0279 | No |
| 90 | S100a13 | 18376 | -0.889 | 0.0017 | No |