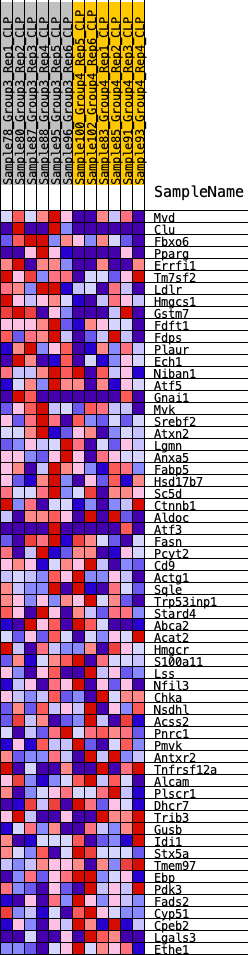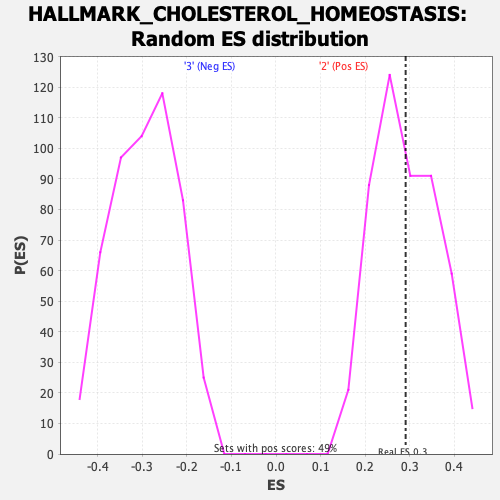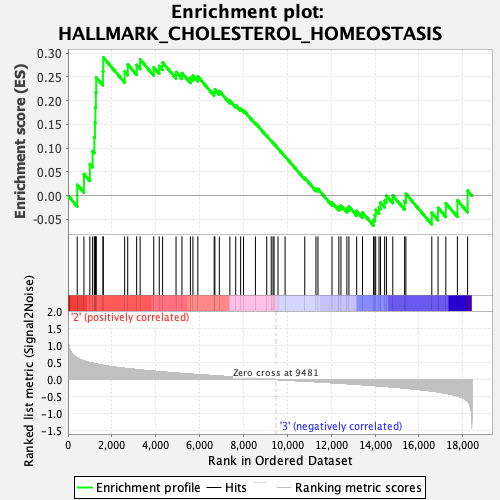
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.29112986 |
| Normalized Enrichment Score (NES) | 1.0013257 |
| Nominal p-value | 0.4601227 |
| FDR q-value | 0.7818899 |
| FWER p-Value | 0.991 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mvd | 420 | 0.635 | 0.0229 | Yes |
| 2 | Clu | 727 | 0.545 | 0.0455 | Yes |
| 3 | Fbxo6 | 996 | 0.491 | 0.0664 | Yes |
| 4 | Pparg | 1121 | 0.475 | 0.0939 | Yes |
| 5 | Errfi1 | 1202 | 0.460 | 0.1227 | Yes |
| 6 | Tm7sf2 | 1226 | 0.457 | 0.1544 | Yes |
| 7 | Ldlr | 1248 | 0.454 | 0.1860 | Yes |
| 8 | Hmgcs1 | 1270 | 0.451 | 0.2174 | Yes |
| 9 | Gstm7 | 1286 | 0.449 | 0.2490 | Yes |
| 10 | Fdft1 | 1592 | 0.414 | 0.2622 | Yes |
| 11 | Fdps | 1608 | 0.412 | 0.2911 | Yes |
| 12 | Plaur | 2583 | 0.325 | 0.2615 | No |
| 13 | Ech1 | 2722 | 0.314 | 0.2766 | No |
| 14 | Niban1 | 3129 | 0.289 | 0.2753 | No |
| 15 | Atf5 | 3288 | 0.278 | 0.2867 | No |
| 16 | Gnai1 | 3909 | 0.241 | 0.2704 | No |
| 17 | Mvk | 4159 | 0.226 | 0.2731 | No |
| 18 | Srebf2 | 4312 | 0.216 | 0.2804 | No |
| 19 | Atxn2 | 4925 | 0.185 | 0.2604 | No |
| 20 | Lgmn | 5195 | 0.173 | 0.2583 | No |
| 21 | Anxa5 | 5580 | 0.154 | 0.2484 | No |
| 22 | Fabp5 | 5694 | 0.149 | 0.2530 | No |
| 23 | Hsd17b7 | 5919 | 0.140 | 0.2510 | No |
| 24 | Sc5d | 6663 | 0.108 | 0.2182 | No |
| 25 | Ctnnb1 | 6696 | 0.107 | 0.2242 | No |
| 26 | Aldoc | 6898 | 0.097 | 0.2203 | No |
| 27 | Atf3 | 7380 | 0.081 | 0.1999 | No |
| 28 | Fasn | 7645 | 0.071 | 0.1906 | No |
| 29 | Pcyt2 | 7870 | 0.062 | 0.1829 | No |
| 30 | Cd9 | 8003 | 0.058 | 0.1799 | No |
| 31 | Actg1 | 8543 | 0.034 | 0.1530 | No |
| 32 | Sqle | 9059 | 0.015 | 0.1260 | No |
| 33 | Trp53inp1 | 9264 | 0.008 | 0.1154 | No |
| 34 | Stard4 | 9346 | 0.004 | 0.1113 | No |
| 35 | Abca2 | 9393 | 0.003 | 0.1090 | No |
| 36 | Acat2 | 9573 | -0.001 | 0.0994 | No |
| 37 | Hmgcr | 9896 | -0.013 | 0.0828 | No |
| 38 | S100a11 | 10793 | -0.046 | 0.0373 | No |
| 39 | Lss | 11303 | -0.064 | 0.0141 | No |
| 40 | Nfil3 | 11395 | -0.067 | 0.0140 | No |
| 41 | Chka | 12034 | -0.092 | -0.0141 | No |
| 42 | Nsdhl | 12347 | -0.104 | -0.0236 | No |
| 43 | Acss2 | 12443 | -0.108 | -0.0210 | No |
| 44 | Pnrc1 | 12713 | -0.120 | -0.0270 | No |
| 45 | Pmvk | 12806 | -0.123 | -0.0231 | No |
| 46 | Antxr2 | 13159 | -0.139 | -0.0322 | No |
| 47 | Tnfrsf12a | 13426 | -0.150 | -0.0359 | No |
| 48 | Alcam | 13928 | -0.174 | -0.0506 | No |
| 49 | Plscr1 | 13986 | -0.177 | -0.0409 | No |
| 50 | Dhcr7 | 14027 | -0.179 | -0.0302 | No |
| 51 | Trib3 | 14177 | -0.187 | -0.0248 | No |
| 52 | Gusb | 14244 | -0.191 | -0.0146 | No |
| 53 | Idi1 | 14434 | -0.201 | -0.0105 | No |
| 54 | Stx5a | 14516 | -0.205 | -0.0001 | No |
| 55 | Tmem97 | 14806 | -0.219 | -0.0001 | No |
| 56 | Ebp | 15344 | -0.249 | -0.0114 | No |
| 57 | Pdk3 | 15402 | -0.253 | 0.0037 | No |
| 58 | Fads2 | 16584 | -0.338 | -0.0363 | No |
| 59 | Cyp51 | 16869 | -0.360 | -0.0258 | No |
| 60 | Cpeb2 | 17223 | -0.402 | -0.0160 | No |
| 61 | Lgals3 | 17751 | -0.483 | -0.0099 | No |
| 62 | Ethe1 | 18216 | -0.633 | 0.0105 | No |