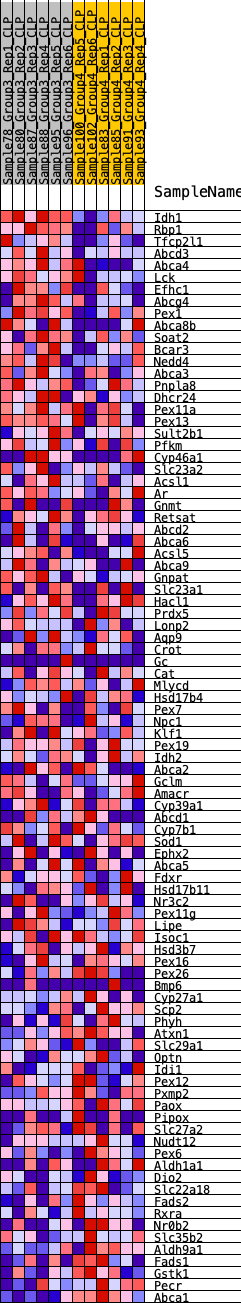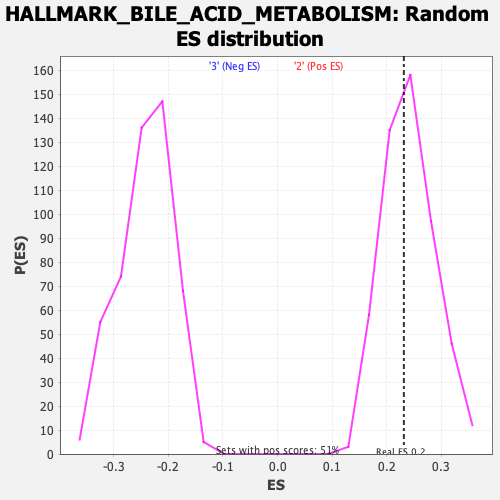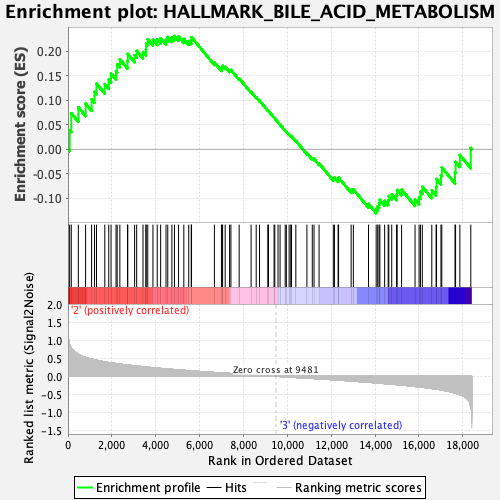
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group3_versus_Group4.Basophil_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | 0.23134981 |
| Normalized Enrichment Score (NES) | 0.9612419 |
| Nominal p-value | 0.5559921 |
| FDR q-value | 0.8435016 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Idh1 | 75 | 0.885 | 0.0390 | Yes |
| 2 | Rbp1 | 149 | 0.787 | 0.0734 | Yes |
| 3 | Tfcp2l1 | 471 | 0.611 | 0.0856 | Yes |
| 4 | Abcd3 | 802 | 0.527 | 0.0933 | Yes |
| 5 | Abca4 | 1076 | 0.482 | 0.1018 | Yes |
| 6 | Lck | 1207 | 0.460 | 0.1171 | Yes |
| 7 | Efhc1 | 1298 | 0.448 | 0.1340 | Yes |
| 8 | Abcg4 | 1677 | 0.402 | 0.1330 | Yes |
| 9 | Pex1 | 1858 | 0.385 | 0.1419 | Yes |
| 10 | Abca8b | 1963 | 0.375 | 0.1544 | Yes |
| 11 | Soat2 | 2190 | 0.355 | 0.1594 | Yes |
| 12 | Bcar3 | 2242 | 0.350 | 0.1737 | Yes |
| 13 | Nedd4 | 2364 | 0.339 | 0.1835 | Yes |
| 14 | Abca3 | 2712 | 0.315 | 0.1799 | Yes |
| 15 | Pnpla8 | 2726 | 0.313 | 0.1945 | Yes |
| 16 | Dhcr24 | 3037 | 0.295 | 0.1919 | Yes |
| 17 | Pex11a | 3131 | 0.289 | 0.2009 | Yes |
| 18 | Pex13 | 3419 | 0.269 | 0.1983 | Yes |
| 19 | Sult2b1 | 3552 | 0.261 | 0.2038 | Yes |
| 20 | Pfkm | 3560 | 0.260 | 0.2161 | Yes |
| 21 | Cyp46a1 | 3636 | 0.255 | 0.2245 | Yes |
| 22 | Slc23a2 | 3872 | 0.244 | 0.2235 | Yes |
| 23 | Acsl1 | 4068 | 0.231 | 0.2241 | Yes |
| 24 | Ar | 4223 | 0.222 | 0.2265 | Yes |
| 25 | Gnmt | 4468 | 0.209 | 0.2234 | Yes |
| 26 | Retsat | 4548 | 0.206 | 0.2291 | Yes |
| 27 | Abcd2 | 4736 | 0.195 | 0.2284 | Yes |
| 28 | Abca6 | 4852 | 0.189 | 0.2313 | Yes |
| 29 | Acsl5 | 5040 | 0.181 | 0.2300 | No |
| 30 | Abca9 | 5280 | 0.169 | 0.2252 | No |
| 31 | Gnpat | 5504 | 0.158 | 0.2207 | No |
| 32 | Slc23a1 | 5617 | 0.153 | 0.2220 | No |
| 33 | Hacl1 | 5631 | 0.152 | 0.2287 | No |
| 34 | Prdx5 | 6674 | 0.108 | 0.1770 | No |
| 35 | Lonp2 | 6994 | 0.094 | 0.1642 | No |
| 36 | Aqp9 | 7022 | 0.092 | 0.1672 | No |
| 37 | Crot | 7048 | 0.091 | 0.1703 | No |
| 38 | Gc | 7165 | 0.087 | 0.1682 | No |
| 39 | Cat | 7365 | 0.082 | 0.1613 | No |
| 40 | Mlycd | 7427 | 0.079 | 0.1618 | No |
| 41 | Hsd17b4 | 7803 | 0.065 | 0.1445 | No |
| 42 | Pex7 | 8346 | 0.044 | 0.1171 | No |
| 43 | Npc1 | 8579 | 0.033 | 0.1060 | No |
| 44 | Klf1 | 8731 | 0.027 | 0.0991 | No |
| 45 | Pex19 | 9109 | 0.013 | 0.0791 | No |
| 46 | Idh2 | 9138 | 0.012 | 0.0782 | No |
| 47 | Abca2 | 9393 | 0.003 | 0.0644 | No |
| 48 | Gclm | 9427 | 0.002 | 0.0627 | No |
| 49 | Amacr | 9574 | -0.001 | 0.0548 | No |
| 50 | Cyp39a1 | 9667 | -0.005 | 0.0500 | No |
| 51 | Abcd1 | 9901 | -0.013 | 0.0379 | No |
| 52 | Cyp7b1 | 9954 | -0.015 | 0.0358 | No |
| 53 | Sod1 | 10080 | -0.019 | 0.0299 | No |
| 54 | Ephx2 | 10155 | -0.022 | 0.0270 | No |
| 55 | Abca5 | 10189 | -0.023 | 0.0263 | No |
| 56 | Fdxr | 10386 | -0.031 | 0.0171 | No |
| 57 | Hsd17b11 | 10891 | -0.050 | -0.0080 | No |
| 58 | Nr3c2 | 11136 | -0.057 | -0.0185 | No |
| 59 | Pex11g | 11216 | -0.061 | -0.0199 | No |
| 60 | Lipe | 11445 | -0.069 | -0.0290 | No |
| 61 | Isoc1 | 12094 | -0.094 | -0.0597 | No |
| 62 | Hsd3b7 | 12145 | -0.096 | -0.0578 | No |
| 63 | Pex16 | 12313 | -0.103 | -0.0619 | No |
| 64 | Pex26 | 12334 | -0.104 | -0.0579 | No |
| 65 | Bmp6 | 12907 | -0.128 | -0.0829 | No |
| 66 | Cyp27a1 | 13008 | -0.132 | -0.0819 | No |
| 67 | Scp2 | 13699 | -0.164 | -0.1116 | No |
| 68 | Phyh | 14048 | -0.181 | -0.1218 | No |
| 69 | Atxn1 | 14118 | -0.184 | -0.1166 | No |
| 70 | Slc29a1 | 14180 | -0.187 | -0.1108 | No |
| 71 | Optn | 14217 | -0.190 | -0.1035 | No |
| 72 | Idi1 | 14434 | -0.201 | -0.1055 | No |
| 73 | Pex12 | 14594 | -0.207 | -0.1041 | No |
| 74 | Pxmp2 | 14625 | -0.209 | -0.0956 | No |
| 75 | Paox | 14757 | -0.216 | -0.0923 | No |
| 76 | Pipox | 14976 | -0.229 | -0.0930 | No |
| 77 | Slc27a2 | 15004 | -0.231 | -0.0832 | No |
| 78 | Nudt12 | 15207 | -0.240 | -0.0826 | No |
| 79 | Pex6 | 15820 | -0.281 | -0.1023 | No |
| 80 | Aldh1a1 | 16004 | -0.293 | -0.0980 | No |
| 81 | Dio2 | 16066 | -0.298 | -0.0868 | No |
| 82 | Slc22a18 | 16151 | -0.304 | -0.0766 | No |
| 83 | Fads2 | 16584 | -0.338 | -0.0837 | No |
| 84 | Rxra | 16780 | -0.353 | -0.0772 | No |
| 85 | Nr0b2 | 16800 | -0.355 | -0.0609 | No |
| 86 | Slc35b2 | 17002 | -0.374 | -0.0537 | No |
| 87 | Aldh9a1 | 17040 | -0.379 | -0.0372 | No |
| 88 | Fads1 | 17643 | -0.465 | -0.0474 | No |
| 89 | Gstk1 | 17668 | -0.470 | -0.0259 | No |
| 90 | Pecr | 17863 | -0.507 | -0.0117 | No |
| 91 | Abca1 | 18362 | -0.851 | 0.0025 | No |