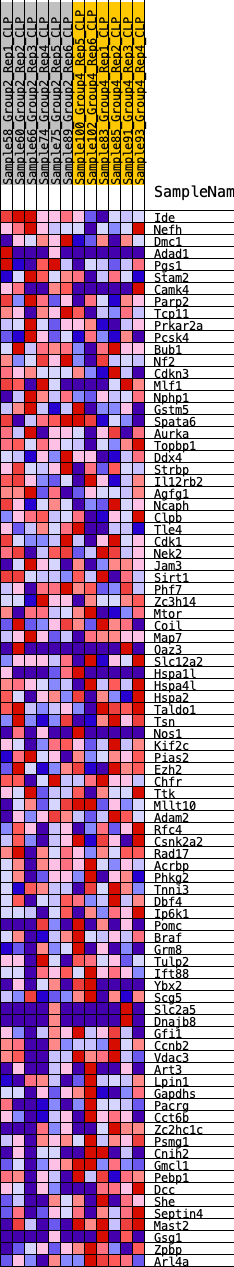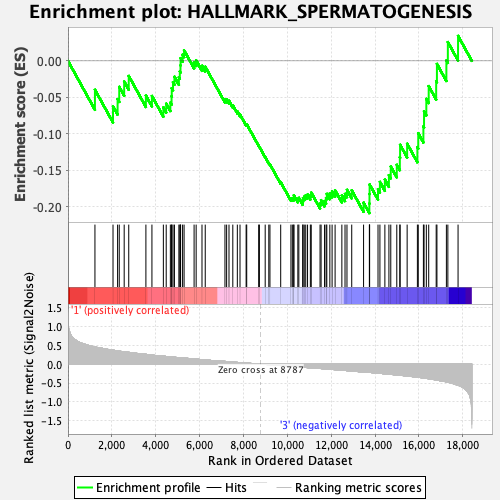
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | -0.20847623 |
| Normalized Enrichment Score (NES) | -0.93116117 |
| Nominal p-value | 0.56063616 |
| FDR q-value | 0.99804026 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ide | 1228 | 0.455 | -0.0395 | No |
| 2 | Nefh | 2052 | 0.368 | -0.0622 | No |
| 3 | Dmc1 | 2261 | 0.351 | -0.0523 | No |
| 4 | Adad1 | 2336 | 0.345 | -0.0355 | No |
| 5 | Pgs1 | 2561 | 0.324 | -0.0282 | No |
| 6 | Stam2 | 2769 | 0.312 | -0.0206 | No |
| 7 | Camk4 | 3551 | 0.260 | -0.0475 | No |
| 8 | Parp2 | 3826 | 0.239 | -0.0480 | No |
| 9 | Tcp11 | 4347 | 0.210 | -0.0637 | No |
| 10 | Prkar2a | 4477 | 0.202 | -0.0585 | No |
| 11 | Pcsk4 | 4666 | 0.193 | -0.0571 | No |
| 12 | Bub1 | 4721 | 0.191 | -0.0485 | No |
| 13 | Nf2 | 4730 | 0.190 | -0.0374 | No |
| 14 | Cdkn3 | 4787 | 0.187 | -0.0292 | No |
| 15 | Mlf1 | 4852 | 0.182 | -0.0217 | No |
| 16 | Nphp1 | 5055 | 0.170 | -0.0225 | No |
| 17 | Gstm5 | 5097 | 0.168 | -0.0146 | No |
| 18 | Spata6 | 5128 | 0.166 | -0.0061 | No |
| 19 | Aurka | 5133 | 0.166 | 0.0037 | No |
| 20 | Topbp1 | 5224 | 0.162 | 0.0085 | No |
| 21 | Ddx4 | 5292 | 0.158 | 0.0145 | No |
| 22 | Strbp | 5748 | 0.136 | -0.0021 | No |
| 23 | Il12rb2 | 5842 | 0.132 | 0.0008 | No |
| 24 | Agfg1 | 6108 | 0.117 | -0.0066 | No |
| 25 | Ncaph | 6256 | 0.110 | -0.0080 | No |
| 26 | Clpb | 7155 | 0.072 | -0.0526 | No |
| 27 | Tle4 | 7231 | 0.069 | -0.0525 | No |
| 28 | Cdk1 | 7339 | 0.063 | -0.0546 | No |
| 29 | Nek2 | 7513 | 0.054 | -0.0607 | No |
| 30 | Jam3 | 7719 | 0.045 | -0.0692 | No |
| 31 | Sirt1 | 7841 | 0.040 | -0.0734 | No |
| 32 | Phf7 | 8120 | 0.028 | -0.0869 | No |
| 33 | Zc3h14 | 8147 | 0.026 | -0.0867 | No |
| 34 | Mtor | 8692 | 0.004 | -0.1162 | No |
| 35 | Coil | 8725 | 0.003 | -0.1178 | No |
| 36 | Map7 | 8989 | -0.007 | -0.1317 | No |
| 37 | Oaz3 | 9154 | -0.014 | -0.1398 | No |
| 38 | Slc12a2 | 9206 | -0.015 | -0.1417 | No |
| 39 | Hspa1l | 9696 | -0.037 | -0.1661 | No |
| 40 | Hspa4l | 10163 | -0.057 | -0.1881 | No |
| 41 | Hspa2 | 10227 | -0.060 | -0.1879 | No |
| 42 | Taldo1 | 10290 | -0.063 | -0.1875 | No |
| 43 | Tsn | 10298 | -0.064 | -0.1840 | No |
| 44 | Nos1 | 10478 | -0.071 | -0.1895 | No |
| 45 | Kif2c | 10523 | -0.073 | -0.1874 | No |
| 46 | Pias2 | 10699 | -0.082 | -0.1920 | No |
| 47 | Ezh2 | 10714 | -0.083 | -0.1878 | No |
| 48 | Chfr | 10769 | -0.085 | -0.1856 | No |
| 49 | Ttk | 10838 | -0.088 | -0.1840 | No |
| 50 | Mllt10 | 10920 | -0.091 | -0.1830 | No |
| 51 | Adam2 | 11048 | -0.097 | -0.1841 | No |
| 52 | Rfc4 | 11088 | -0.099 | -0.1802 | No |
| 53 | Csnk2a2 | 11490 | -0.114 | -0.1952 | Yes |
| 54 | Rad17 | 11537 | -0.116 | -0.1907 | Yes |
| 55 | Acrbp | 11695 | -0.123 | -0.1918 | Yes |
| 56 | Phkg2 | 11764 | -0.126 | -0.1879 | Yes |
| 57 | Tnni3 | 11798 | -0.128 | -0.1820 | Yes |
| 58 | Dbf4 | 11936 | -0.134 | -0.1814 | Yes |
| 59 | Ip6k1 | 12039 | -0.139 | -0.1785 | Yes |
| 60 | Pomc | 12176 | -0.145 | -0.1772 | Yes |
| 61 | Braf | 12484 | -0.160 | -0.1843 | Yes |
| 62 | Grm8 | 12628 | -0.166 | -0.1820 | Yes |
| 63 | Tulp2 | 12717 | -0.171 | -0.1765 | Yes |
| 64 | Ift88 | 12934 | -0.183 | -0.1773 | Yes |
| 65 | Ybx2 | 13474 | -0.208 | -0.1941 | Yes |
| 66 | Scg5 | 13738 | -0.219 | -0.1952 | Yes |
| 67 | Slc2a5 | 13744 | -0.219 | -0.1822 | Yes |
| 68 | Dnajb8 | 13750 | -0.219 | -0.1693 | Yes |
| 69 | Gfi1 | 14132 | -0.238 | -0.1756 | Yes |
| 70 | Ccnb2 | 14219 | -0.243 | -0.1657 | Yes |
| 71 | Vdac3 | 14445 | -0.255 | -0.1625 | Yes |
| 72 | Art3 | 14629 | -0.266 | -0.1564 | Yes |
| 73 | Lpin1 | 14714 | -0.271 | -0.1447 | Yes |
| 74 | Gapdhs | 14988 | -0.286 | -0.1423 | Yes |
| 75 | Pacrg | 15127 | -0.293 | -0.1320 | Yes |
| 76 | Cct6b | 15139 | -0.294 | -0.1149 | Yes |
| 77 | Zc2hc1c | 15460 | -0.313 | -0.1134 | Yes |
| 78 | Psmg1 | 15927 | -0.345 | -0.1180 | Yes |
| 79 | Cnih2 | 15967 | -0.348 | -0.0991 | Yes |
| 80 | Gmcl1 | 16201 | -0.366 | -0.0897 | Yes |
| 81 | Pebp1 | 16231 | -0.368 | -0.0690 | Yes |
| 82 | Dcc | 16335 | -0.375 | -0.0520 | Yes |
| 83 | She | 16443 | -0.386 | -0.0345 | Yes |
| 84 | Septin4 | 16784 | -0.418 | -0.0278 | Yes |
| 85 | Mast2 | 16818 | -0.421 | -0.0041 | Yes |
| 86 | Gsg1 | 17249 | -0.467 | 0.0006 | Yes |
| 87 | Zpbp | 17316 | -0.475 | 0.0258 | Yes |
| 88 | Arl4a | 17781 | -0.559 | 0.0342 | Yes |