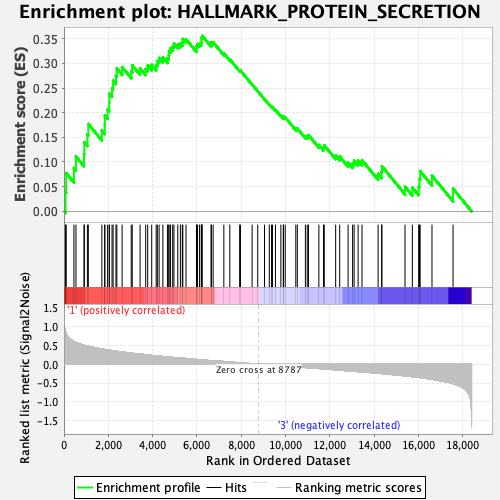
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.3558865 |
| Normalized Enrichment Score (NES) | 1.5873816 |
| Nominal p-value | 0.01039501 |
| FDR q-value | 0.33069247 |
| FWER p-Value | 0.289 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Napg | 69 | 0.874 | 0.0394 | Yes |
| 2 | Gla | 100 | 0.800 | 0.0773 | Yes |
| 3 | Mon2 | 448 | 0.598 | 0.0878 | Yes |
| 4 | Tpd52 | 540 | 0.571 | 0.1111 | Yes |
| 5 | Cltc | 904 | 0.501 | 0.1160 | Yes |
| 6 | Rab5a | 918 | 0.498 | 0.1399 | Yes |
| 7 | Stx16 | 1057 | 0.479 | 0.1560 | Yes |
| 8 | Arfgef2 | 1098 | 0.472 | 0.1771 | Yes |
| 9 | Rab22a | 1710 | 0.402 | 0.1636 | Yes |
| 10 | Atp1a1 | 1839 | 0.387 | 0.1758 | Yes |
| 11 | Rps6ka3 | 1844 | 0.387 | 0.1946 | Yes |
| 12 | Igf2r | 1970 | 0.376 | 0.2064 | Yes |
| 13 | M6pr | 2044 | 0.369 | 0.2206 | Yes |
| 14 | Cln5 | 2047 | 0.368 | 0.2387 | Yes |
| 15 | Atp6v1h | 2169 | 0.361 | 0.2499 | Yes |
| 16 | Pam | 2217 | 0.356 | 0.2649 | Yes |
| 17 | Copb1 | 2346 | 0.344 | 0.2749 | Yes |
| 18 | Egfr | 2387 | 0.339 | 0.2895 | Yes |
| 19 | Arcn1 | 2624 | 0.322 | 0.2925 | Yes |
| 20 | Tspan8 | 3036 | 0.291 | 0.2844 | Yes |
| 21 | Tmed10 | 3082 | 0.288 | 0.2962 | Yes |
| 22 | Atp7a | 3435 | 0.266 | 0.2901 | Yes |
| 23 | Tmx1 | 3689 | 0.249 | 0.2887 | Yes |
| 24 | Sec31a | 3780 | 0.242 | 0.2957 | Yes |
| 25 | Copb2 | 3961 | 0.229 | 0.2972 | Yes |
| 26 | Snap23 | 4163 | 0.220 | 0.2971 | Yes |
| 27 | Ap2b1 | 4215 | 0.217 | 0.3050 | Yes |
| 28 | Vamp3 | 4296 | 0.213 | 0.3111 | Yes |
| 29 | Rab9 | 4465 | 0.203 | 0.3119 | Yes |
| 30 | Cope | 4671 | 0.193 | 0.3103 | Yes |
| 31 | Vps45 | 4732 | 0.190 | 0.3164 | Yes |
| 32 | Ap2m1 | 4744 | 0.190 | 0.3252 | Yes |
| 33 | Golga4 | 4812 | 0.185 | 0.3306 | Yes |
| 34 | Uso1 | 4908 | 0.179 | 0.3343 | Yes |
| 35 | Anp32e | 4964 | 0.175 | 0.3399 | Yes |
| 36 | Ergic3 | 5138 | 0.166 | 0.3387 | Yes |
| 37 | Rab14 | 5256 | 0.160 | 0.3402 | Yes |
| 38 | Clta | 5343 | 0.155 | 0.3432 | Yes |
| 39 | Ocrl | 5366 | 0.154 | 0.3496 | Yes |
| 40 | Sec22b | 5510 | 0.148 | 0.3491 | Yes |
| 41 | Adam10 | 5982 | 0.125 | 0.3296 | Yes |
| 42 | Mapk1 | 5990 | 0.124 | 0.3353 | Yes |
| 43 | Scamp3 | 6029 | 0.122 | 0.3393 | Yes |
| 44 | Bnip3 | 6121 | 0.117 | 0.3401 | Yes |
| 45 | Cog2 | 6199 | 0.113 | 0.3415 | Yes |
| 46 | Snx2 | 6210 | 0.113 | 0.3465 | Yes |
| 47 | Arfgef1 | 6211 | 0.113 | 0.3521 | Yes |
| 48 | Cav2 | 6243 | 0.111 | 0.3559 | Yes |
| 49 | Vps4b | 6643 | 0.095 | 0.3388 | No |
| 50 | Arfip1 | 6654 | 0.094 | 0.3429 | No |
| 51 | Ctsc | 6735 | 0.090 | 0.3430 | No |
| 52 | Kif1b | 7218 | 0.070 | 0.3201 | No |
| 53 | Stx7 | 7488 | 0.055 | 0.3082 | No |
| 54 | Lman1 | 7932 | 0.036 | 0.2858 | No |
| 55 | Clcn3 | 7969 | 0.034 | 0.2855 | No |
| 56 | Rab2a | 8495 | 0.012 | 0.2574 | No |
| 57 | Ap3b1 | 8749 | 0.001 | 0.2436 | No |
| 58 | Sec24d | 9055 | -0.010 | 0.2275 | No |
| 59 | Rer1 | 9271 | -0.018 | 0.2166 | No |
| 60 | Lamp2 | 9373 | -0.023 | 0.2123 | No |
| 61 | Vamp4 | 9414 | -0.025 | 0.2113 | No |
| 62 | Dst | 9548 | -0.030 | 0.2055 | No |
| 63 | Gosr2 | 9793 | -0.041 | 0.1942 | No |
| 64 | Scamp1 | 9906 | -0.046 | 0.1904 | No |
| 65 | Yipf6 | 9908 | -0.046 | 0.1926 | No |
| 66 | Galc | 9990 | -0.050 | 0.1906 | No |
| 67 | Arfgap3 | 10468 | -0.071 | 0.1681 | No |
| 68 | Ap1g1 | 10536 | -0.074 | 0.1681 | No |
| 69 | Gnas | 10901 | -0.090 | 0.1526 | No |
| 70 | Sh3gl2 | 10985 | -0.094 | 0.1528 | No |
| 71 | Tom1l1 | 11040 | -0.096 | 0.1546 | No |
| 72 | Arf1 | 11510 | -0.115 | 0.1346 | No |
| 73 | Abca1 | 11718 | -0.124 | 0.1295 | No |
| 74 | Zw10 | 11753 | -0.125 | 0.1338 | No |
| 75 | Ppt1 | 12268 | -0.149 | 0.1131 | No |
| 76 | Dop1a | 12451 | -0.158 | 0.1110 | No |
| 77 | Stx12 | 12830 | -0.177 | 0.0991 | No |
| 78 | Gbf1 | 13033 | -0.187 | 0.0973 | No |
| 79 | Bet1 | 13102 | -0.190 | 0.1029 | No |
| 80 | Cd63 | 13282 | -0.200 | 0.1030 | No |
| 81 | Stam | 13457 | -0.207 | 0.1037 | No |
| 82 | Ap2s1 | 14185 | -0.240 | 0.0759 | No |
| 83 | Ykt6 | 14345 | -0.250 | 0.0796 | No |
| 84 | Ica1 | 14356 | -0.251 | 0.0914 | No |
| 85 | Tsg101 | 15400 | -0.310 | 0.0498 | No |
| 86 | Krt18 | 15731 | -0.331 | 0.0481 | No |
| 87 | Dnm1l | 16015 | -0.352 | 0.0501 | No |
| 88 | Sgms1 | 16051 | -0.353 | 0.0656 | No |
| 89 | Napa | 16084 | -0.356 | 0.0814 | No |
| 90 | Sod1 | 16618 | -0.402 | 0.0722 | No |
| 91 | Ap3s1 | 17569 | -0.516 | 0.0458 | No |