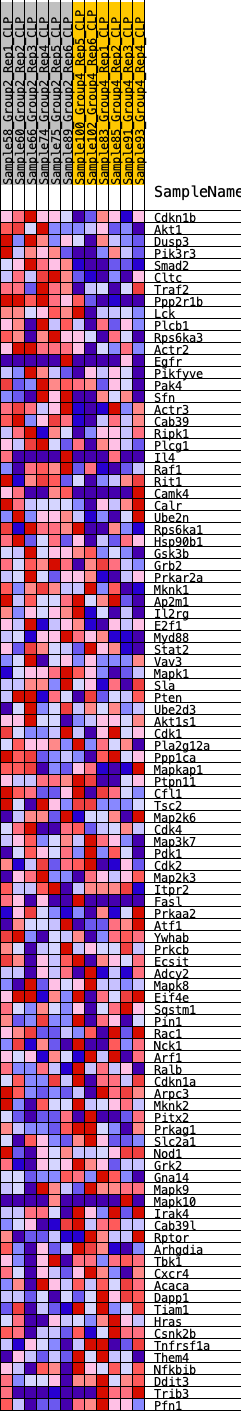
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
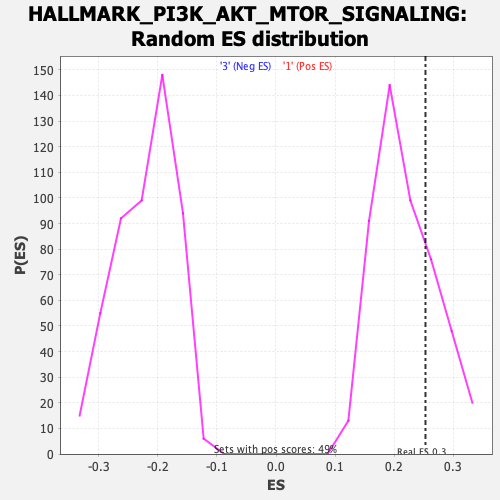
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.25330204 |
| Normalized Enrichment Score (NES) | 1.1599799 |
| Nominal p-value | 0.25050917 |
| FDR q-value | 0.92021656 |
| FWER p-Value | 0.96 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cdkn1b | 528 | 0.575 | -0.0031 | Yes |
| 2 | Akt1 | 592 | 0.560 | 0.0185 | Yes |
| 3 | Dusp3 | 648 | 0.549 | 0.0401 | Yes |
| 4 | Pik3r3 | 687 | 0.541 | 0.0623 | Yes |
| 5 | Smad2 | 824 | 0.516 | 0.0779 | Yes |
| 6 | Cltc | 904 | 0.501 | 0.0961 | Yes |
| 7 | Traf2 | 1080 | 0.475 | 0.1078 | Yes |
| 8 | Ppp2r1b | 1232 | 0.454 | 0.1199 | Yes |
| 9 | Lck | 1395 | 0.434 | 0.1304 | Yes |
| 10 | Plcb1 | 1838 | 0.388 | 0.1236 | Yes |
| 11 | Rps6ka3 | 1844 | 0.387 | 0.1407 | Yes |
| 12 | Actr2 | 2078 | 0.366 | 0.1443 | Yes |
| 13 | Egfr | 2387 | 0.339 | 0.1427 | Yes |
| 14 | Pikfyve | 2412 | 0.337 | 0.1565 | Yes |
| 15 | Pak4 | 2427 | 0.335 | 0.1707 | Yes |
| 16 | Sfn | 2473 | 0.330 | 0.1830 | Yes |
| 17 | Actr3 | 2631 | 0.321 | 0.1888 | Yes |
| 18 | Cab39 | 2846 | 0.305 | 0.1908 | Yes |
| 19 | Ripk1 | 2876 | 0.303 | 0.2028 | Yes |
| 20 | Plcg1 | 3004 | 0.294 | 0.2090 | Yes |
| 21 | Il4 | 3093 | 0.288 | 0.2170 | Yes |
| 22 | Raf1 | 3515 | 0.262 | 0.2058 | Yes |
| 23 | Rit1 | 3535 | 0.261 | 0.2165 | Yes |
| 24 | Camk4 | 3551 | 0.260 | 0.2273 | Yes |
| 25 | Calr | 3777 | 0.242 | 0.2259 | Yes |
| 26 | Ube2n | 3818 | 0.240 | 0.2344 | Yes |
| 27 | Rps6ka1 | 3870 | 0.235 | 0.2421 | Yes |
| 28 | Hsp90b1 | 4030 | 0.225 | 0.2435 | Yes |
| 29 | Gsk3b | 4037 | 0.225 | 0.2533 | Yes |
| 30 | Grb2 | 4395 | 0.207 | 0.2431 | No |
| 31 | Prkar2a | 4477 | 0.202 | 0.2477 | No |
| 32 | Mknk1 | 4613 | 0.195 | 0.2491 | No |
| 33 | Ap2m1 | 4744 | 0.190 | 0.2505 | No |
| 34 | Il2rg | 4924 | 0.177 | 0.2486 | No |
| 35 | E2f1 | 5053 | 0.170 | 0.2493 | No |
| 36 | Myd88 | 5355 | 0.155 | 0.2398 | No |
| 37 | Stat2 | 5459 | 0.151 | 0.2409 | No |
| 38 | Vav3 | 5789 | 0.134 | 0.2289 | No |
| 39 | Mapk1 | 5990 | 0.124 | 0.2236 | No |
| 40 | Sla | 6021 | 0.123 | 0.2274 | No |
| 41 | Pten | 6248 | 0.111 | 0.2201 | No |
| 42 | Ube2d3 | 6776 | 0.089 | 0.1953 | No |
| 43 | Akt1s1 | 7070 | 0.075 | 0.1826 | No |
| 44 | Cdk1 | 7339 | 0.063 | 0.1708 | No |
| 45 | Pla2g12a | 7360 | 0.062 | 0.1725 | No |
| 46 | Ppp1ca | 7467 | 0.057 | 0.1692 | No |
| 47 | Mapkap1 | 7724 | 0.045 | 0.1573 | No |
| 48 | Ptpn11 | 7794 | 0.041 | 0.1554 | No |
| 49 | Cfl1 | 7949 | 0.035 | 0.1485 | No |
| 50 | Tsc2 | 8106 | 0.028 | 0.1412 | No |
| 51 | Map2k6 | 8220 | 0.023 | 0.1361 | No |
| 52 | Cdk4 | 8258 | 0.022 | 0.1350 | No |
| 53 | Map3k7 | 8263 | 0.022 | 0.1358 | No |
| 54 | Pdk1 | 8318 | 0.019 | 0.1337 | No |
| 55 | Cdk2 | 8389 | 0.016 | 0.1306 | No |
| 56 | Map2k3 | 8398 | 0.016 | 0.1308 | No |
| 57 | Itpr2 | 8458 | 0.014 | 0.1282 | No |
| 58 | Fasl | 8554 | 0.009 | 0.1235 | No |
| 59 | Prkaa2 | 9038 | -0.009 | 0.0975 | No |
| 60 | Atf1 | 9195 | -0.015 | 0.0896 | No |
| 61 | Ywhab | 9298 | -0.020 | 0.0850 | No |
| 62 | Prkcb | 9756 | -0.039 | 0.0618 | No |
| 63 | Ecsit | 9854 | -0.044 | 0.0584 | No |
| 64 | Adcy2 | 10171 | -0.057 | 0.0437 | No |
| 65 | Mapk8 | 10385 | -0.067 | 0.0351 | No |
| 66 | Eif4e | 10449 | -0.070 | 0.0348 | No |
| 67 | Sqstm1 | 10772 | -0.085 | 0.0210 | No |
| 68 | Pin1 | 10915 | -0.090 | 0.0173 | No |
| 69 | Rac1 | 10978 | -0.094 | 0.0181 | No |
| 70 | Nck1 | 11244 | -0.105 | 0.0083 | No |
| 71 | Arf1 | 11510 | -0.115 | -0.0010 | No |
| 72 | Ralb | 11518 | -0.115 | 0.0038 | No |
| 73 | Cdkn1a | 11830 | -0.130 | -0.0074 | No |
| 74 | Arpc3 | 11836 | -0.130 | -0.0019 | No |
| 75 | Mknk2 | 12143 | -0.144 | -0.0122 | No |
| 76 | Pitx2 | 12229 | -0.148 | -0.0102 | No |
| 77 | Prkag1 | 12506 | -0.161 | -0.0180 | No |
| 78 | Slc2a1 | 12813 | -0.176 | -0.0269 | No |
| 79 | Nod1 | 13300 | -0.200 | -0.0444 | No |
| 80 | Grk2 | 13428 | -0.205 | -0.0422 | No |
| 81 | Gna14 | 13541 | -0.211 | -0.0388 | No |
| 82 | Mapk9 | 13664 | -0.216 | -0.0358 | No |
| 83 | Mapk10 | 13748 | -0.219 | -0.0305 | No |
| 84 | Irak4 | 13807 | -0.221 | -0.0238 | No |
| 85 | Cab39l | 13855 | -0.224 | -0.0164 | No |
| 86 | Rptor | 14278 | -0.246 | -0.0284 | No |
| 87 | Arhgdia | 14307 | -0.248 | -0.0188 | No |
| 88 | Tbk1 | 14478 | -0.257 | -0.0166 | No |
| 89 | Cxcr4 | 14718 | -0.271 | -0.0175 | No |
| 90 | Acaca | 14962 | -0.284 | -0.0181 | No |
| 91 | Dapp1 | 15485 | -0.315 | -0.0325 | No |
| 92 | Tiam1 | 15670 | -0.328 | -0.0279 | No |
| 93 | Hras | 16054 | -0.354 | -0.0330 | No |
| 94 | Csnk2b | 16288 | -0.372 | -0.0290 | No |
| 95 | Tnfrsf1a | 17257 | -0.468 | -0.0610 | No |
| 96 | Them4 | 17388 | -0.486 | -0.0463 | No |
| 97 | Nfkbib | 17692 | -0.543 | -0.0385 | No |
| 98 | Ddit3 | 17713 | -0.547 | -0.0151 | No |
| 99 | Trib3 | 17838 | -0.570 | 0.0036 | No |
| 100 | Pfn1 | 17982 | -0.614 | 0.0233 | No |