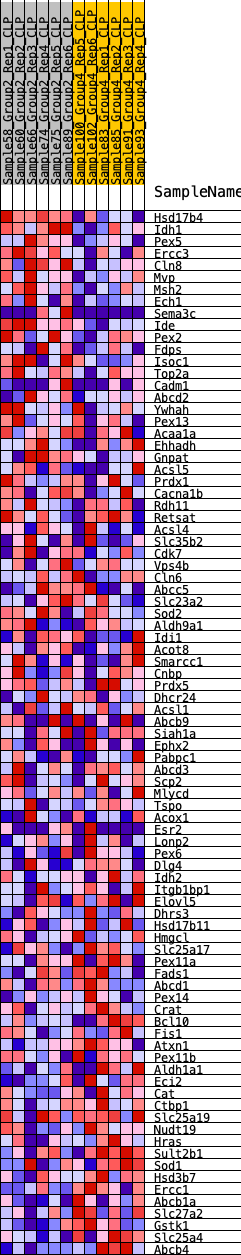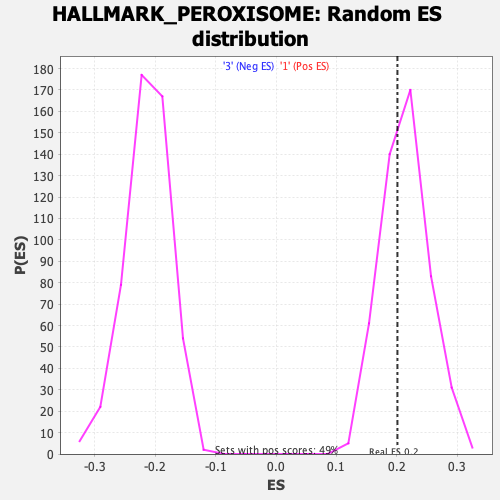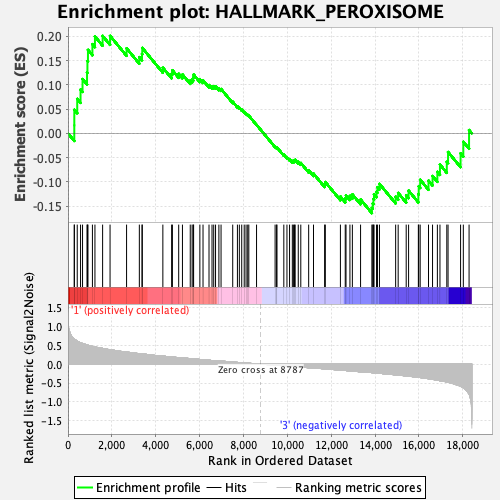
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.2008386 |
| Normalized Enrichment Score (NES) | 0.93918943 |
| Nominal p-value | 0.61054766 |
| FDR q-value | 0.977171 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsd17b4 | 287 | 0.662 | 0.0165 | Yes |
| 2 | Idh1 | 288 | 0.662 | 0.0487 | Yes |
| 3 | Pex5 | 421 | 0.607 | 0.0710 | Yes |
| 4 | Ercc3 | 572 | 0.565 | 0.0903 | Yes |
| 5 | Cln8 | 663 | 0.545 | 0.1119 | Yes |
| 6 | Mvp | 868 | 0.508 | 0.1254 | Yes |
| 7 | Msh2 | 886 | 0.504 | 0.1490 | Yes |
| 8 | Ech1 | 910 | 0.499 | 0.1720 | Yes |
| 9 | Sema3c | 1113 | 0.469 | 0.1837 | Yes |
| 10 | Ide | 1228 | 0.455 | 0.1997 | Yes |
| 11 | Pex2 | 1578 | 0.415 | 0.2008 | Yes |
| 12 | Fdps | 1917 | 0.381 | 0.2008 | Yes |
| 13 | Isoc1 | 2670 | 0.317 | 0.1752 | No |
| 14 | Top2a | 3252 | 0.278 | 0.1570 | No |
| 15 | Cadm1 | 3374 | 0.271 | 0.1635 | No |
| 16 | Abcd2 | 3386 | 0.270 | 0.1760 | No |
| 17 | Ywhah | 4324 | 0.212 | 0.1352 | No |
| 18 | Pex13 | 4724 | 0.191 | 0.1227 | No |
| 19 | Acaa1a | 4759 | 0.188 | 0.1300 | No |
| 20 | Ehhadh | 5046 | 0.171 | 0.1227 | No |
| 21 | Gnpat | 5217 | 0.162 | 0.1213 | No |
| 22 | Acsl5 | 5573 | 0.145 | 0.1089 | No |
| 23 | Prdx1 | 5658 | 0.142 | 0.1112 | No |
| 24 | Cacna1b | 5711 | 0.138 | 0.1151 | No |
| 25 | Rdh11 | 5726 | 0.138 | 0.1211 | No |
| 26 | Retsat | 6011 | 0.123 | 0.1116 | No |
| 27 | Acsl4 | 6155 | 0.115 | 0.1094 | No |
| 28 | Slc35b2 | 6428 | 0.102 | 0.0995 | No |
| 29 | Cdk7 | 6565 | 0.095 | 0.0967 | No |
| 30 | Vps4b | 6643 | 0.095 | 0.0971 | No |
| 31 | Cln6 | 6726 | 0.091 | 0.0970 | No |
| 32 | Abcc5 | 6878 | 0.084 | 0.0929 | No |
| 33 | Slc23a2 | 6977 | 0.079 | 0.0914 | No |
| 34 | Sod2 | 7509 | 0.055 | 0.0651 | No |
| 35 | Aldh9a1 | 7723 | 0.045 | 0.0556 | No |
| 36 | Idi1 | 7811 | 0.041 | 0.0528 | No |
| 37 | Acot8 | 7919 | 0.036 | 0.0488 | No |
| 38 | Smarcc1 | 8033 | 0.031 | 0.0441 | No |
| 39 | Cnbp | 8117 | 0.028 | 0.0409 | No |
| 40 | Prdx5 | 8193 | 0.024 | 0.0380 | No |
| 41 | Dhcr24 | 8243 | 0.022 | 0.0364 | No |
| 42 | Acsl1 | 8595 | 0.007 | 0.0176 | No |
| 43 | Abcb9 | 9441 | -0.026 | -0.0272 | No |
| 44 | Siah1a | 9509 | -0.028 | -0.0295 | No |
| 45 | Ephx2 | 9525 | -0.029 | -0.0289 | No |
| 46 | Pabpc1 | 9838 | -0.043 | -0.0439 | No |
| 47 | Abcd3 | 9981 | -0.049 | -0.0492 | No |
| 48 | Scp2 | 10100 | -0.054 | -0.0530 | No |
| 49 | Mlycd | 10226 | -0.060 | -0.0569 | No |
| 50 | Tspo | 10275 | -0.063 | -0.0565 | No |
| 51 | Acox1 | 10326 | -0.065 | -0.0560 | No |
| 52 | Esr2 | 10355 | -0.066 | -0.0544 | No |
| 53 | Lonp2 | 10497 | -0.072 | -0.0586 | No |
| 54 | Pex6 | 10612 | -0.078 | -0.0610 | No |
| 55 | Dlg4 | 10971 | -0.093 | -0.0760 | No |
| 56 | Idh2 | 11191 | -0.102 | -0.0830 | No |
| 57 | Itgb1bp1 | 11698 | -0.123 | -0.1046 | No |
| 58 | Elovl5 | 11730 | -0.124 | -0.1003 | No |
| 59 | Dhrs3 | 12418 | -0.157 | -0.1301 | No |
| 60 | Hsd17b11 | 12638 | -0.167 | -0.1340 | No |
| 61 | Hmgcl | 12673 | -0.169 | -0.1276 | No |
| 62 | Slc25a17 | 12852 | -0.178 | -0.1287 | No |
| 63 | Pex11a | 12963 | -0.184 | -0.1257 | No |
| 64 | Fads1 | 13337 | -0.202 | -0.1362 | No |
| 65 | Abcd1 | 13853 | -0.223 | -0.1535 | No |
| 66 | Pex14 | 13892 | -0.226 | -0.1446 | No |
| 67 | Crat | 13923 | -0.228 | -0.1351 | No |
| 68 | Bcl10 | 13950 | -0.229 | -0.1254 | No |
| 69 | Fis1 | 14065 | -0.235 | -0.1202 | No |
| 70 | Atxn1 | 14107 | -0.237 | -0.1109 | No |
| 71 | Pex11b | 14198 | -0.241 | -0.1041 | No |
| 72 | Aldh1a1 | 14936 | -0.282 | -0.1306 | No |
| 73 | Eci2 | 15051 | -0.290 | -0.1227 | No |
| 74 | Cat | 15417 | -0.311 | -0.1275 | No |
| 75 | Ctbp1 | 15526 | -0.318 | -0.1180 | No |
| 76 | Slc25a19 | 15971 | -0.348 | -0.1253 | No |
| 77 | Nudt19 | 15988 | -0.350 | -0.1092 | No |
| 78 | Hras | 16054 | -0.354 | -0.0955 | No |
| 79 | Sult2b1 | 16434 | -0.385 | -0.0975 | No |
| 80 | Sod1 | 16618 | -0.402 | -0.0879 | No |
| 81 | Hsd3b7 | 16838 | -0.423 | -0.0793 | No |
| 82 | Ercc1 | 16956 | -0.438 | -0.0644 | No |
| 83 | Abcb1a | 17268 | -0.469 | -0.0586 | No |
| 84 | Slc27a2 | 17325 | -0.477 | -0.0384 | No |
| 85 | Gstk1 | 17896 | -0.585 | -0.0411 | No |
| 86 | Slc25a4 | 18021 | -0.625 | -0.0175 | No |
| 87 | Abcb4 | 18283 | -0.793 | 0.0068 | No |