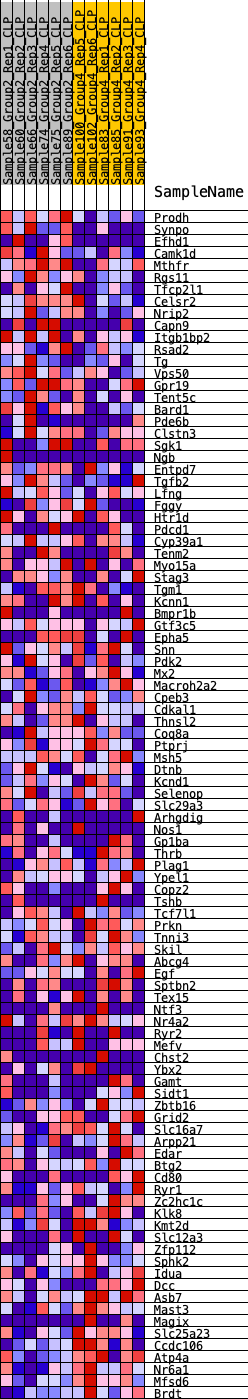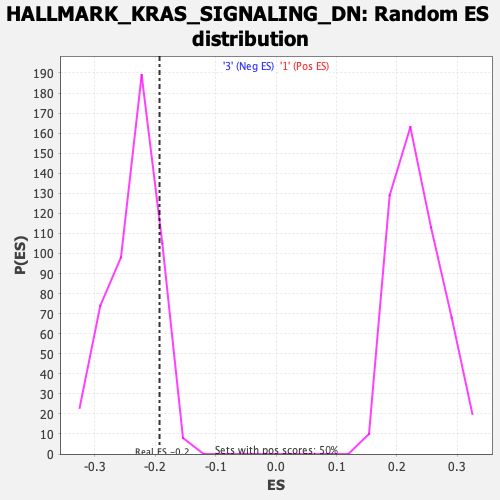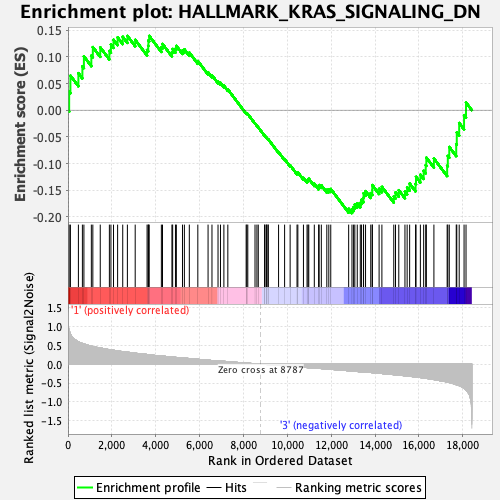
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.19302085 |
| Normalized Enrichment Score (NES) | -0.8192869 |
| Nominal p-value | 0.87525153 |
| FDR q-value | 0.95591646 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prodh | 53 | 0.899 | 0.0347 | No |
| 2 | Synpo | 110 | 0.791 | 0.0647 | No |
| 3 | Efhd1 | 474 | 0.585 | 0.0694 | No |
| 4 | Camk1d | 656 | 0.547 | 0.0823 | No |
| 5 | Mthfr | 725 | 0.532 | 0.1009 | No |
| 6 | Rgs11 | 1064 | 0.478 | 0.1024 | No |
| 7 | Tfcp2l1 | 1132 | 0.467 | 0.1183 | No |
| 8 | Celsr2 | 1470 | 0.427 | 0.1177 | No |
| 9 | Nrip2 | 1886 | 0.383 | 0.1111 | No |
| 10 | Capn9 | 1957 | 0.378 | 0.1230 | No |
| 11 | Itgb1bp2 | 2075 | 0.366 | 0.1320 | No |
| 12 | Rsad2 | 2262 | 0.351 | 0.1365 | No |
| 13 | Tg | 2495 | 0.329 | 0.1376 | No |
| 14 | Vps50 | 2707 | 0.314 | 0.1392 | No |
| 15 | Gpr19 | 3063 | 0.289 | 0.1319 | No |
| 16 | Tent5c | 3605 | 0.255 | 0.1130 | No |
| 17 | Bard1 | 3657 | 0.252 | 0.1208 | No |
| 18 | Pde6b | 3666 | 0.251 | 0.1308 | No |
| 19 | Clstn3 | 3702 | 0.248 | 0.1393 | No |
| 20 | Sgk1 | 4256 | 0.214 | 0.1181 | No |
| 21 | Ngb | 4308 | 0.212 | 0.1241 | No |
| 22 | Entpd7 | 4738 | 0.190 | 0.1087 | No |
| 23 | Tgfb2 | 4771 | 0.187 | 0.1147 | No |
| 24 | Lfng | 4899 | 0.180 | 0.1153 | No |
| 25 | Fggy | 4939 | 0.177 | 0.1206 | No |
| 26 | Htr1d | 5219 | 0.162 | 0.1121 | No |
| 27 | Pdcd1 | 5305 | 0.158 | 0.1141 | No |
| 28 | Cyp39a1 | 5530 | 0.147 | 0.1080 | No |
| 29 | Tenm2 | 5918 | 0.128 | 0.0922 | No |
| 30 | Myo15a | 6385 | 0.104 | 0.0711 | No |
| 31 | Stag3 | 6563 | 0.095 | 0.0654 | No |
| 32 | Tgm1 | 6839 | 0.086 | 0.0540 | No |
| 33 | Kcnn1 | 6951 | 0.080 | 0.0513 | No |
| 34 | Bmpr1b | 7106 | 0.074 | 0.0460 | No |
| 35 | Gtf3c5 | 7286 | 0.066 | 0.0390 | No |
| 36 | Epha5 | 8121 | 0.028 | -0.0054 | No |
| 37 | Snn | 8151 | 0.026 | -0.0059 | No |
| 38 | Pdk2 | 8196 | 0.024 | -0.0073 | No |
| 39 | Mx2 | 8525 | 0.010 | -0.0248 | No |
| 40 | Macroh2a2 | 8612 | 0.007 | -0.0292 | No |
| 41 | Cpeb3 | 8674 | 0.005 | -0.0323 | No |
| 42 | Cdkal1 | 8960 | -0.006 | -0.0477 | No |
| 43 | Thnsl2 | 8978 | -0.006 | -0.0483 | No |
| 44 | Coq8a | 9042 | -0.009 | -0.0514 | No |
| 45 | Ptprj | 9056 | -0.010 | -0.0517 | No |
| 46 | Msh5 | 9117 | -0.013 | -0.0544 | No |
| 47 | Dtnb | 9128 | -0.013 | -0.0544 | No |
| 48 | Kcnd1 | 9599 | -0.032 | -0.0788 | No |
| 49 | Selenop | 9874 | -0.044 | -0.0919 | No |
| 50 | Slc29a3 | 10126 | -0.055 | -0.1033 | No |
| 51 | Arhgdig | 10440 | -0.069 | -0.1175 | No |
| 52 | Nos1 | 10478 | -0.071 | -0.1165 | No |
| 53 | Gp1ba | 10733 | -0.083 | -0.1269 | No |
| 54 | Thrb | 10897 | -0.090 | -0.1320 | No |
| 55 | Plag1 | 10942 | -0.092 | -0.1306 | No |
| 56 | Ypel1 | 10976 | -0.094 | -0.1285 | No |
| 57 | Copz2 | 11228 | -0.104 | -0.1378 | No |
| 58 | Tshb | 11425 | -0.112 | -0.1439 | No |
| 59 | Tcf7l1 | 11447 | -0.112 | -0.1403 | No |
| 60 | Prkn | 11548 | -0.117 | -0.1409 | No |
| 61 | Tnni3 | 11798 | -0.128 | -0.1492 | No |
| 62 | Skil | 11889 | -0.132 | -0.1485 | No |
| 63 | Abcg4 | 11978 | -0.136 | -0.1476 | No |
| 64 | Egf | 12794 | -0.175 | -0.1848 | No |
| 65 | Sptbn2 | 12945 | -0.183 | -0.1854 | Yes |
| 66 | Tex15 | 13019 | -0.186 | -0.1816 | Yes |
| 67 | Ntf3 | 13075 | -0.189 | -0.1767 | Yes |
| 68 | Nr4a2 | 13186 | -0.194 | -0.1746 | Yes |
| 69 | Ryr2 | 13324 | -0.201 | -0.1736 | Yes |
| 70 | Mefv | 13376 | -0.204 | -0.1679 | Yes |
| 71 | Chst2 | 13468 | -0.207 | -0.1642 | Yes |
| 72 | Ybx2 | 13474 | -0.208 | -0.1558 | Yes |
| 73 | Gamt | 13564 | -0.212 | -0.1518 | Yes |
| 74 | Sidt1 | 13801 | -0.221 | -0.1554 | Yes |
| 75 | Zbtb16 | 13871 | -0.225 | -0.1498 | Yes |
| 76 | Grid2 | 13872 | -0.225 | -0.1404 | Yes |
| 77 | Slc16a7 | 14182 | -0.240 | -0.1472 | Yes |
| 78 | Arpp21 | 14309 | -0.248 | -0.1437 | Yes |
| 79 | Edar | 14851 | -0.278 | -0.1616 | Yes |
| 80 | Btg2 | 14930 | -0.282 | -0.1541 | Yes |
| 81 | Cd80 | 15079 | -0.291 | -0.1500 | Yes |
| 82 | Ryr1 | 15360 | -0.307 | -0.1525 | Yes |
| 83 | Zc2hc1c | 15460 | -0.313 | -0.1448 | Yes |
| 84 | Klk8 | 15574 | -0.321 | -0.1375 | Yes |
| 85 | Kmt2d | 15844 | -0.339 | -0.1381 | Yes |
| 86 | Slc12a3 | 15864 | -0.340 | -0.1249 | Yes |
| 87 | Zfp112 | 16063 | -0.355 | -0.1208 | Yes |
| 88 | Sphk2 | 16213 | -0.367 | -0.1136 | Yes |
| 89 | Idua | 16307 | -0.373 | -0.1031 | Yes |
| 90 | Dcc | 16335 | -0.375 | -0.0889 | Yes |
| 91 | Asb7 | 16680 | -0.408 | -0.0907 | Yes |
| 92 | Mast3 | 17284 | -0.470 | -0.1039 | Yes |
| 93 | Magix | 17314 | -0.475 | -0.0856 | Yes |
| 94 | Slc25a23 | 17381 | -0.485 | -0.0690 | Yes |
| 95 | Ccdc106 | 17698 | -0.544 | -0.0635 | Yes |
| 96 | Atp4a | 17723 | -0.549 | -0.0418 | Yes |
| 97 | Nr6a1 | 17833 | -0.569 | -0.0240 | Yes |
| 98 | Mfsd6 | 18055 | -0.639 | -0.0093 | Yes |
| 99 | Brdt | 18144 | -0.683 | 0.0144 | Yes |