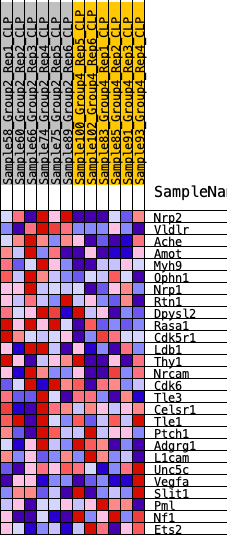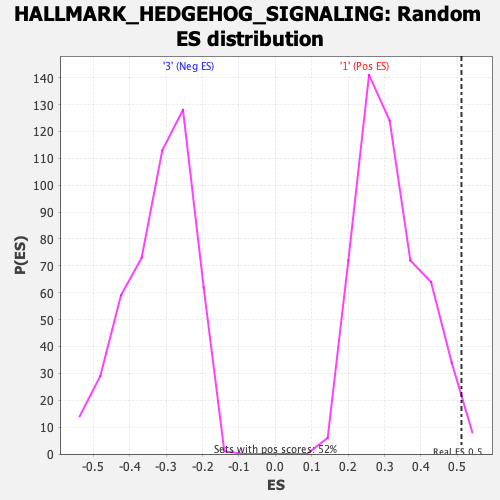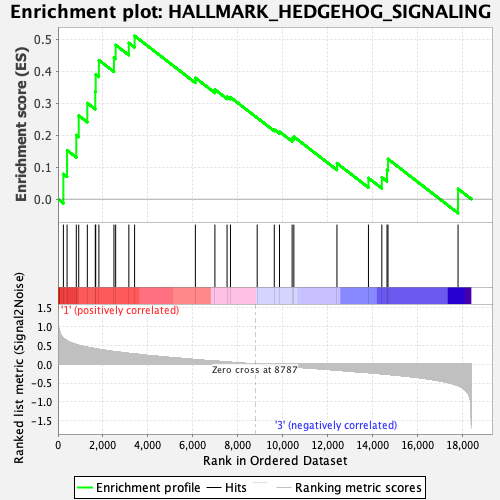
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.5112192 |
| Normalized Enrichment Score (NES) | 1.6101825 |
| Nominal p-value | 0.015355086 |
| FDR q-value | 0.5243579 |
| FWER p-Value | 0.232 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nrp2 | 240 | 0.679 | 0.0787 | Yes |
| 2 | Vldlr | 404 | 0.616 | 0.1529 | Yes |
| 3 | Ache | 815 | 0.517 | 0.2004 | Yes |
| 4 | Amot | 923 | 0.496 | 0.2616 | Yes |
| 5 | Myh9 | 1306 | 0.445 | 0.3009 | Yes |
| 6 | Ophn1 | 1660 | 0.407 | 0.3366 | Yes |
| 7 | Nrp1 | 1679 | 0.405 | 0.3902 | Yes |
| 8 | Rtn1 | 1821 | 0.389 | 0.4350 | Yes |
| 9 | Dpysl2 | 2492 | 0.330 | 0.4431 | Yes |
| 10 | Rasa1 | 2568 | 0.324 | 0.4827 | Yes |
| 11 | Cdk5r1 | 3155 | 0.283 | 0.4890 | Yes |
| 12 | Ldb1 | 3411 | 0.268 | 0.5112 | Yes |
| 13 | Thy1 | 6118 | 0.117 | 0.3798 | No |
| 14 | Nrcam | 6988 | 0.079 | 0.3432 | No |
| 15 | Cdk6 | 7529 | 0.054 | 0.3210 | No |
| 16 | Tle3 | 7681 | 0.046 | 0.3191 | No |
| 17 | Celsr1 | 8871 | -0.001 | 0.2546 | No |
| 18 | Tle1 | 9632 | -0.034 | 0.2178 | No |
| 19 | Ptch1 | 9864 | -0.044 | 0.2111 | No |
| 20 | Adgrg1 | 10427 | -0.069 | 0.1899 | No |
| 21 | L1cam | 10504 | -0.072 | 0.1955 | No |
| 22 | Unc5c | 12423 | -0.157 | 0.1123 | No |
| 23 | Vegfa | 13826 | -0.222 | 0.0661 | No |
| 24 | Slit1 | 14422 | -0.254 | 0.0680 | No |
| 25 | Pml | 14653 | -0.268 | 0.0916 | No |
| 26 | Nf1 | 14696 | -0.269 | 0.1257 | No |
| 27 | Ets2 | 17816 | -0.564 | 0.0322 | No |