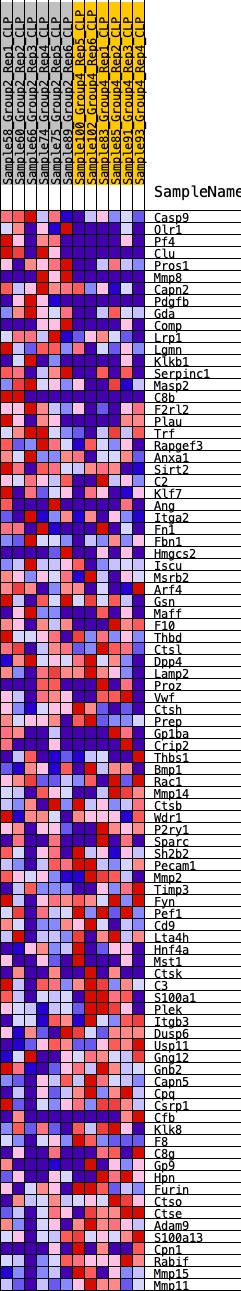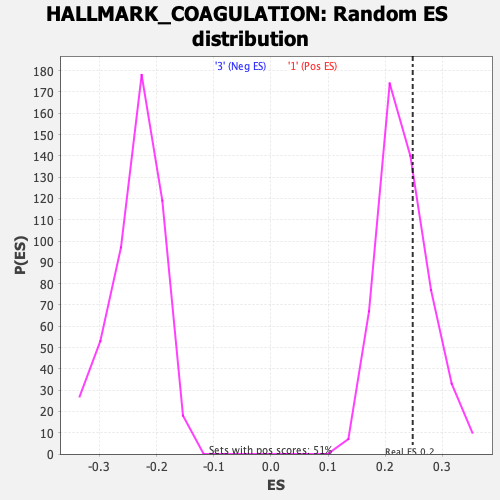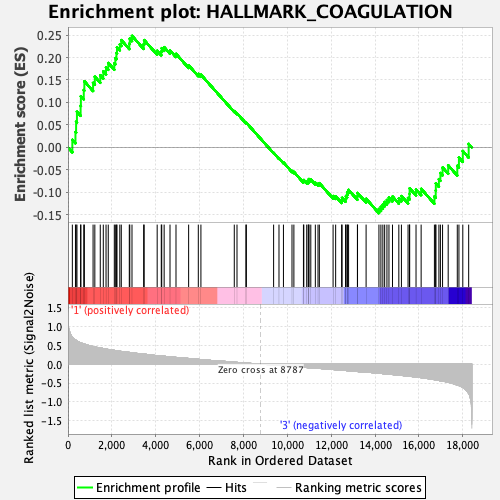
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.24809505 |
| Normalized Enrichment Score (NES) | 1.0669603 |
| Nominal p-value | 0.3051181 |
| FDR q-value | 0.9389134 |
| FWER p-Value | 0.995 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Casp9 | 198 | 0.709 | 0.0170 | Yes |
| 2 | Olr1 | 340 | 0.639 | 0.0344 | Yes |
| 3 | Pf4 | 370 | 0.629 | 0.0575 | Yes |
| 4 | Clu | 408 | 0.612 | 0.0795 | Yes |
| 5 | Pros1 | 575 | 0.564 | 0.0925 | Yes |
| 6 | Mmp8 | 587 | 0.561 | 0.1139 | Yes |
| 7 | Capn2 | 719 | 0.533 | 0.1277 | Yes |
| 8 | Pdgfb | 746 | 0.530 | 0.1470 | Yes |
| 9 | Gda | 1143 | 0.465 | 0.1437 | Yes |
| 10 | Comp | 1223 | 0.456 | 0.1572 | Yes |
| 11 | Lrp1 | 1471 | 0.427 | 0.1605 | Yes |
| 12 | Lgmn | 1609 | 0.412 | 0.1691 | Yes |
| 13 | Klkb1 | 1734 | 0.399 | 0.1780 | Yes |
| 14 | Serpinc1 | 1835 | 0.388 | 0.1878 | Yes |
| 15 | Masp2 | 2111 | 0.364 | 0.1870 | Yes |
| 16 | C8b | 2157 | 0.361 | 0.1988 | Yes |
| 17 | F2rl2 | 2207 | 0.357 | 0.2101 | Yes |
| 18 | Plau | 2233 | 0.355 | 0.2226 | Yes |
| 19 | Trf | 2362 | 0.342 | 0.2291 | Yes |
| 20 | Rapgef3 | 2429 | 0.335 | 0.2386 | Yes |
| 21 | Anxa1 | 2794 | 0.310 | 0.2309 | Yes |
| 22 | Sirt2 | 2816 | 0.307 | 0.2418 | Yes |
| 23 | C2 | 2917 | 0.299 | 0.2481 | Yes |
| 24 | Klf7 | 3449 | 0.265 | 0.2295 | No |
| 25 | Ang | 3472 | 0.264 | 0.2386 | No |
| 26 | Itga2 | 4065 | 0.224 | 0.2151 | No |
| 27 | Fn1 | 4252 | 0.215 | 0.2134 | No |
| 28 | Fbn1 | 4273 | 0.214 | 0.2207 | No |
| 29 | Hmgcs2 | 4383 | 0.208 | 0.2229 | No |
| 30 | Iscu | 4650 | 0.194 | 0.2160 | No |
| 31 | Msrb2 | 4923 | 0.178 | 0.2081 | No |
| 32 | Arf4 | 5499 | 0.149 | 0.1825 | No |
| 33 | Gsn | 5940 | 0.127 | 0.1635 | No |
| 34 | Maff | 6059 | 0.120 | 0.1617 | No |
| 35 | F10 | 7580 | 0.051 | 0.0808 | No |
| 36 | Thbd | 7704 | 0.046 | 0.0759 | No |
| 37 | Ctsl | 8109 | 0.028 | 0.0549 | No |
| 38 | Dpp4 | 8131 | 0.027 | 0.0548 | No |
| 39 | Lamp2 | 9373 | -0.023 | -0.0120 | No |
| 40 | Proz | 9618 | -0.033 | -0.0240 | No |
| 41 | Vwf | 9821 | -0.042 | -0.0334 | No |
| 42 | Ctsh | 10211 | -0.059 | -0.0523 | No |
| 43 | Prep | 10297 | -0.064 | -0.0545 | No |
| 44 | Gp1ba | 10733 | -0.083 | -0.0750 | No |
| 45 | Crip2 | 10749 | -0.084 | -0.0725 | No |
| 46 | Thbs1 | 10882 | -0.089 | -0.0762 | No |
| 47 | Bmp1 | 10949 | -0.092 | -0.0762 | No |
| 48 | Rac1 | 10978 | -0.094 | -0.0740 | No |
| 49 | Mmp14 | 10979 | -0.094 | -0.0703 | No |
| 50 | Ctsb | 11068 | -0.098 | -0.0713 | No |
| 51 | Wdr1 | 11275 | -0.105 | -0.0784 | No |
| 52 | P2ry1 | 11397 | -0.111 | -0.0807 | No |
| 53 | Sparc | 11458 | -0.113 | -0.0795 | No |
| 54 | Sh2b2 | 12084 | -0.141 | -0.1081 | No |
| 55 | Pecam1 | 12201 | -0.146 | -0.1087 | No |
| 56 | Mmp2 | 12474 | -0.159 | -0.1173 | No |
| 57 | Timp3 | 12493 | -0.161 | -0.1120 | No |
| 58 | Fyn | 12653 | -0.168 | -0.1141 | No |
| 59 | Pef1 | 12670 | -0.169 | -0.1083 | No |
| 60 | Cd9 | 12723 | -0.171 | -0.1045 | No |
| 61 | Lta4h | 12746 | -0.173 | -0.0989 | No |
| 62 | Hnf4a | 12791 | -0.175 | -0.0944 | No |
| 63 | Mst1 | 13193 | -0.194 | -0.1087 | No |
| 64 | Ctsk | 13198 | -0.195 | -0.1013 | No |
| 65 | C3 | 13589 | -0.213 | -0.1142 | No |
| 66 | S100a1 | 14174 | -0.240 | -0.1367 | No |
| 67 | Plek | 14264 | -0.245 | -0.1319 | No |
| 68 | Itgb3 | 14353 | -0.251 | -0.1269 | No |
| 69 | Dusp6 | 14435 | -0.254 | -0.1213 | No |
| 70 | Usp11 | 14539 | -0.261 | -0.1167 | No |
| 71 | Gng12 | 14631 | -0.267 | -0.1112 | No |
| 72 | Gnb2 | 14792 | -0.274 | -0.1092 | No |
| 73 | Capn5 | 15081 | -0.291 | -0.1135 | No |
| 74 | Cpq | 15199 | -0.298 | -0.1082 | No |
| 75 | Csrp1 | 15502 | -0.316 | -0.1123 | No |
| 76 | Cfb | 15570 | -0.320 | -0.1034 | No |
| 77 | Klk8 | 15574 | -0.321 | -0.0910 | No |
| 78 | F8 | 15865 | -0.341 | -0.0934 | No |
| 79 | C8g | 16098 | -0.357 | -0.0921 | No |
| 80 | Gp9 | 16703 | -0.410 | -0.1090 | No |
| 81 | Hpn | 16762 | -0.415 | -0.0959 | No |
| 82 | Furin | 16768 | -0.416 | -0.0798 | No |
| 83 | Ctso | 16908 | -0.431 | -0.0705 | No |
| 84 | Ctse | 16976 | -0.440 | -0.0569 | No |
| 85 | Adam9 | 17075 | -0.449 | -0.0446 | No |
| 86 | S100a13 | 17331 | -0.477 | -0.0398 | No |
| 87 | Cpn1 | 17746 | -0.553 | -0.0407 | No |
| 88 | Rabif | 17821 | -0.567 | -0.0225 | No |
| 89 | Mmp15 | 17997 | -0.616 | -0.0079 | No |
| 90 | Mmp11 | 18265 | -0.772 | 0.0078 | No |