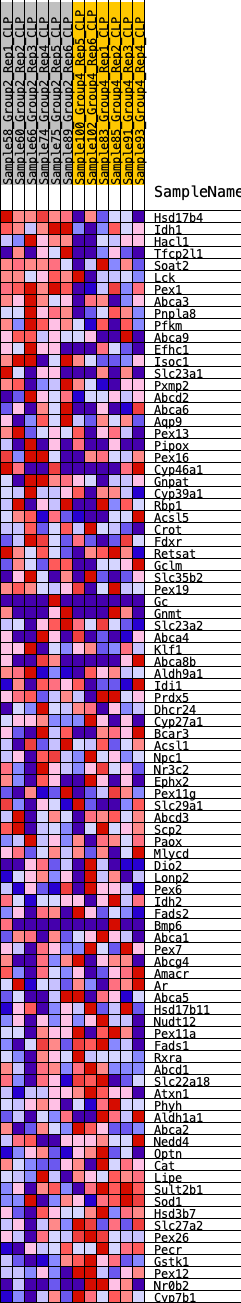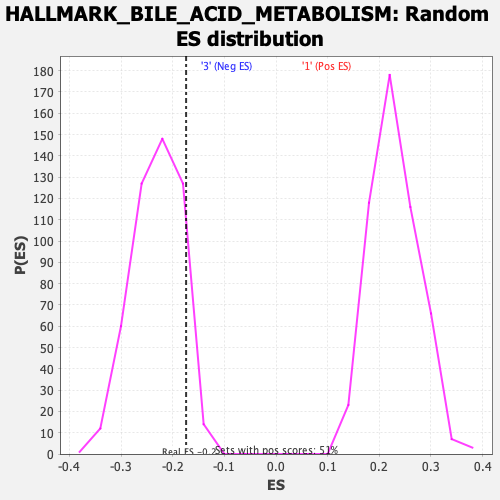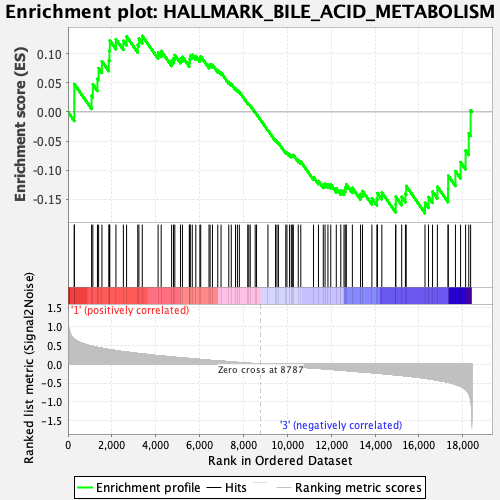
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group4.Basophil_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.17406192 |
| Normalized Enrichment Score (NES) | -0.75401247 |
| Nominal p-value | 0.90593046 |
| FDR q-value | 0.9033376 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hsd17b4 | 287 | 0.662 | 0.0162 | No |
| 2 | Idh1 | 288 | 0.662 | 0.0480 | No |
| 3 | Hacl1 | 1076 | 0.476 | 0.0280 | No |
| 4 | Tfcp2l1 | 1132 | 0.467 | 0.0475 | No |
| 5 | Soat2 | 1342 | 0.440 | 0.0572 | No |
| 6 | Lck | 1395 | 0.434 | 0.0753 | No |
| 7 | Pex1 | 1547 | 0.419 | 0.0872 | No |
| 8 | Abca3 | 1860 | 0.385 | 0.0887 | No |
| 9 | Pnpla8 | 1889 | 0.383 | 0.1056 | No |
| 10 | Pfkm | 1906 | 0.381 | 0.1231 | No |
| 11 | Abca9 | 2186 | 0.359 | 0.1251 | No |
| 12 | Efhc1 | 2524 | 0.327 | 0.1225 | No |
| 13 | Isoc1 | 2670 | 0.317 | 0.1298 | No |
| 14 | Slc23a1 | 3180 | 0.281 | 0.1156 | No |
| 15 | Pxmp2 | 3232 | 0.279 | 0.1262 | No |
| 16 | Abcd2 | 3386 | 0.270 | 0.1308 | No |
| 17 | Abca6 | 4107 | 0.222 | 0.1022 | No |
| 18 | Aqp9 | 4248 | 0.215 | 0.1049 | No |
| 19 | Pex13 | 4724 | 0.191 | 0.0881 | No |
| 20 | Pipox | 4811 | 0.185 | 0.0923 | No |
| 21 | Pex16 | 4869 | 0.181 | 0.0979 | No |
| 22 | Cyp46a1 | 5127 | 0.166 | 0.0919 | No |
| 23 | Gnpat | 5217 | 0.162 | 0.0948 | No |
| 24 | Cyp39a1 | 5530 | 0.147 | 0.0849 | No |
| 25 | Rbp1 | 5553 | 0.146 | 0.0907 | No |
| 26 | Acsl5 | 5573 | 0.145 | 0.0967 | No |
| 27 | Crot | 5665 | 0.141 | 0.0985 | No |
| 28 | Fdxr | 5826 | 0.132 | 0.0961 | No |
| 29 | Retsat | 6011 | 0.123 | 0.0920 | No |
| 30 | Gclm | 6050 | 0.121 | 0.0958 | No |
| 31 | Slc35b2 | 6428 | 0.102 | 0.0801 | No |
| 32 | Pex19 | 6478 | 0.099 | 0.0822 | No |
| 33 | Gc | 6585 | 0.095 | 0.0810 | No |
| 34 | Gnmt | 6827 | 0.086 | 0.0720 | No |
| 35 | Slc23a2 | 6977 | 0.079 | 0.0677 | No |
| 36 | Abca4 | 7329 | 0.063 | 0.0516 | No |
| 37 | Klf1 | 7443 | 0.058 | 0.0482 | No |
| 38 | Abca8b | 7636 | 0.049 | 0.0400 | No |
| 39 | Aldh9a1 | 7723 | 0.045 | 0.0375 | No |
| 40 | Idi1 | 7811 | 0.041 | 0.0347 | No |
| 41 | Prdx5 | 8193 | 0.024 | 0.0151 | No |
| 42 | Dhcr24 | 8243 | 0.022 | 0.0135 | No |
| 43 | Cyp27a1 | 8307 | 0.020 | 0.0110 | No |
| 44 | Bcar3 | 8533 | 0.010 | -0.0008 | No |
| 45 | Acsl1 | 8595 | 0.007 | -0.0038 | No |
| 46 | Npc1 | 9115 | -0.013 | -0.0315 | No |
| 47 | Nr3c2 | 9461 | -0.027 | -0.0491 | No |
| 48 | Ephx2 | 9525 | -0.029 | -0.0511 | No |
| 49 | Pex11g | 9592 | -0.032 | -0.0532 | No |
| 50 | Slc29a1 | 9928 | -0.047 | -0.0692 | No |
| 51 | Abcd3 | 9981 | -0.049 | -0.0697 | No |
| 52 | Scp2 | 10100 | -0.054 | -0.0735 | No |
| 53 | Paox | 10180 | -0.058 | -0.0750 | No |
| 54 | Mlycd | 10226 | -0.060 | -0.0746 | No |
| 55 | Dio2 | 10272 | -0.063 | -0.0740 | No |
| 56 | Lonp2 | 10497 | -0.072 | -0.0828 | No |
| 57 | Pex6 | 10612 | -0.078 | -0.0853 | No |
| 58 | Idh2 | 11191 | -0.102 | -0.1119 | No |
| 59 | Fads2 | 11417 | -0.111 | -0.1188 | No |
| 60 | Bmp6 | 11637 | -0.120 | -0.1250 | No |
| 61 | Abca1 | 11718 | -0.124 | -0.1234 | No |
| 62 | Pex7 | 11850 | -0.131 | -0.1243 | No |
| 63 | Abcg4 | 11978 | -0.136 | -0.1246 | No |
| 64 | Amacr | 12230 | -0.148 | -0.1312 | No |
| 65 | Ar | 12435 | -0.158 | -0.1348 | No |
| 66 | Abca5 | 12583 | -0.164 | -0.1349 | No |
| 67 | Hsd17b11 | 12638 | -0.167 | -0.1298 | No |
| 68 | Nudt12 | 12688 | -0.170 | -0.1243 | No |
| 69 | Pex11a | 12963 | -0.184 | -0.1304 | No |
| 70 | Fads1 | 13337 | -0.202 | -0.1411 | No |
| 71 | Rxra | 13425 | -0.205 | -0.1359 | No |
| 72 | Abcd1 | 13853 | -0.223 | -0.1485 | No |
| 73 | Slc22a18 | 14082 | -0.236 | -0.1496 | No |
| 74 | Atxn1 | 14107 | -0.237 | -0.1395 | No |
| 75 | Phyh | 14306 | -0.248 | -0.1384 | No |
| 76 | Aldh1a1 | 14936 | -0.282 | -0.1591 | Yes |
| 77 | Abca2 | 14943 | -0.283 | -0.1458 | Yes |
| 78 | Nedd4 | 15212 | -0.299 | -0.1461 | Yes |
| 79 | Optn | 15382 | -0.308 | -0.1405 | Yes |
| 80 | Cat | 15417 | -0.311 | -0.1273 | Yes |
| 81 | Lipe | 16274 | -0.371 | -0.1562 | Yes |
| 82 | Sult2b1 | 16434 | -0.385 | -0.1463 | Yes |
| 83 | Sod1 | 16618 | -0.402 | -0.1370 | Yes |
| 84 | Hsd3b7 | 16838 | -0.423 | -0.1286 | Yes |
| 85 | Slc27a2 | 17325 | -0.477 | -0.1321 | Yes |
| 86 | Pex26 | 17332 | -0.477 | -0.1095 | Yes |
| 87 | Pecr | 17662 | -0.534 | -0.1017 | Yes |
| 88 | Gstk1 | 17896 | -0.585 | -0.0863 | Yes |
| 89 | Pex12 | 18125 | -0.674 | -0.0663 | Yes |
| 90 | Nr0b2 | 18269 | -0.775 | -0.0368 | Yes |
| 91 | Cyp7b1 | 18354 | -0.921 | 0.0029 | Yes |