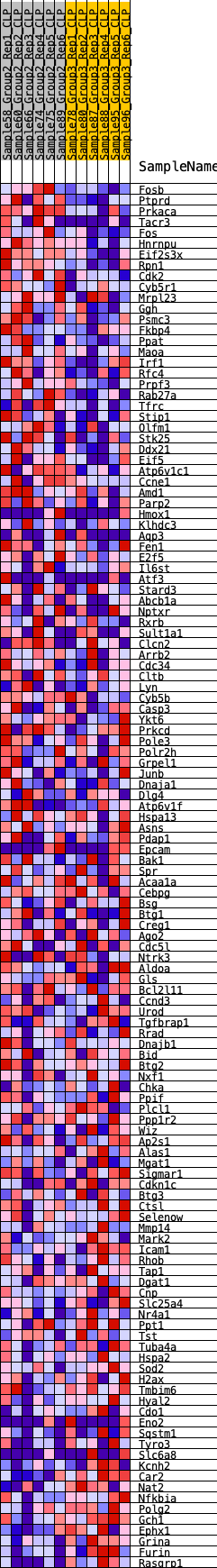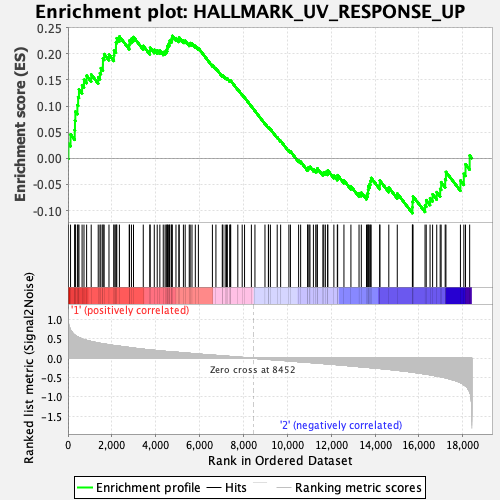
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | 0.23429197 |
| Normalized Enrichment Score (NES) | 0.9146016 |
| Nominal p-value | 0.5907336 |
| FDR q-value | 0.8841242 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fosb | 20 | 0.928 | 0.0280 | Yes |
| 2 | Ptprd | 116 | 0.729 | 0.0457 | Yes |
| 3 | Prkaca | 307 | 0.597 | 0.0540 | Yes |
| 4 | Tacr3 | 318 | 0.592 | 0.0721 | Yes |
| 5 | Fos | 335 | 0.582 | 0.0894 | Yes |
| 6 | Hnrnpu | 428 | 0.557 | 0.1019 | Yes |
| 7 | Eif2s3x | 461 | 0.547 | 0.1173 | Yes |
| 8 | Rpn1 | 500 | 0.535 | 0.1320 | Yes |
| 9 | Cdk2 | 642 | 0.497 | 0.1399 | Yes |
| 10 | Cyb5r1 | 729 | 0.480 | 0.1503 | Yes |
| 11 | Mrpl23 | 849 | 0.461 | 0.1582 | Yes |
| 12 | Ggh | 1058 | 0.432 | 0.1604 | Yes |
| 13 | Psmc3 | 1378 | 0.392 | 0.1552 | Yes |
| 14 | Fkbp4 | 1457 | 0.384 | 0.1630 | Yes |
| 15 | Ppat | 1500 | 0.380 | 0.1726 | Yes |
| 16 | Maoa | 1592 | 0.371 | 0.1793 | Yes |
| 17 | Irf1 | 1593 | 0.371 | 0.1909 | Yes |
| 18 | Rfc4 | 1648 | 0.366 | 0.1995 | Yes |
| 19 | Prpf3 | 1868 | 0.347 | 0.1984 | Yes |
| 20 | Rab27a | 2091 | 0.328 | 0.1965 | Yes |
| 21 | Tfrc | 2100 | 0.327 | 0.2063 | Yes |
| 22 | Stip1 | 2186 | 0.321 | 0.2118 | Yes |
| 23 | Olfm1 | 2187 | 0.321 | 0.2219 | Yes |
| 24 | Stk25 | 2224 | 0.318 | 0.2299 | Yes |
| 25 | Ddx21 | 2343 | 0.310 | 0.2332 | Yes |
| 26 | Eif5 | 2793 | 0.276 | 0.2172 | Yes |
| 27 | Atp6v1c1 | 2801 | 0.275 | 0.2255 | Yes |
| 28 | Ccne1 | 2892 | 0.269 | 0.2290 | Yes |
| 29 | Amd1 | 2986 | 0.261 | 0.2321 | Yes |
| 30 | Parp2 | 3431 | 0.233 | 0.2151 | Yes |
| 31 | Hmox1 | 3733 | 0.215 | 0.2054 | Yes |
| 32 | Klhdc3 | 3742 | 0.214 | 0.2117 | Yes |
| 33 | Aqp3 | 3933 | 0.204 | 0.2077 | Yes |
| 34 | Fen1 | 4067 | 0.196 | 0.2066 | Yes |
| 35 | E2f5 | 4187 | 0.189 | 0.2060 | Yes |
| 36 | Il6st | 4342 | 0.181 | 0.2033 | Yes |
| 37 | Atf3 | 4426 | 0.177 | 0.2043 | Yes |
| 38 | Stard3 | 4481 | 0.174 | 0.2068 | Yes |
| 39 | Abcb1a | 4522 | 0.172 | 0.2100 | Yes |
| 40 | Nptxr | 4532 | 0.172 | 0.2149 | Yes |
| 41 | Rxrb | 4574 | 0.170 | 0.2179 | Yes |
| 42 | Sult1a1 | 4623 | 0.168 | 0.2206 | Yes |
| 43 | Clcn2 | 4638 | 0.167 | 0.2251 | Yes |
| 44 | Arrb2 | 4711 | 0.163 | 0.2262 | Yes |
| 45 | Cdc34 | 4738 | 0.162 | 0.2299 | Yes |
| 46 | Cltb | 4751 | 0.161 | 0.2343 | Yes |
| 47 | Lyn | 4922 | 0.155 | 0.2299 | No |
| 48 | Cyb5b | 5056 | 0.149 | 0.2272 | No |
| 49 | Casp3 | 5067 | 0.148 | 0.2313 | No |
| 50 | Ykt6 | 5261 | 0.139 | 0.2251 | No |
| 51 | Prkcd | 5342 | 0.134 | 0.2250 | No |
| 52 | Pole3 | 5527 | 0.126 | 0.2189 | No |
| 53 | Polr2h | 5569 | 0.125 | 0.2205 | No |
| 54 | Grpel1 | 5652 | 0.120 | 0.2198 | No |
| 55 | Junb | 5804 | 0.113 | 0.2151 | No |
| 56 | Dnaja1 | 5944 | 0.107 | 0.2109 | No |
| 57 | Dlg4 | 6583 | 0.079 | 0.1784 | No |
| 58 | Atp6v1f | 6743 | 0.072 | 0.1720 | No |
| 59 | Hspa13 | 7033 | 0.061 | 0.1581 | No |
| 60 | Asns | 7061 | 0.060 | 0.1585 | No |
| 61 | Pdap1 | 7165 | 0.055 | 0.1546 | No |
| 62 | Epcam | 7225 | 0.053 | 0.1530 | No |
| 63 | Bak1 | 7264 | 0.051 | 0.1526 | No |
| 64 | Spr | 7371 | 0.045 | 0.1482 | No |
| 65 | Acaa1a | 7402 | 0.044 | 0.1479 | No |
| 66 | Cebpg | 7417 | 0.043 | 0.1485 | No |
| 67 | Bsg | 7741 | 0.029 | 0.1317 | No |
| 68 | Btg1 | 7941 | 0.022 | 0.1215 | No |
| 69 | Creg1 | 8048 | 0.016 | 0.1162 | No |
| 70 | Ago2 | 8360 | 0.004 | 0.0993 | No |
| 71 | Cdc5l | 8522 | -0.002 | 0.0906 | No |
| 72 | Ntrk3 | 8979 | -0.021 | 0.0663 | No |
| 73 | Aldoa | 9135 | -0.028 | 0.0587 | No |
| 74 | Gls | 9141 | -0.028 | 0.0593 | No |
| 75 | Bcl2l11 | 9226 | -0.032 | 0.0557 | No |
| 76 | Ccnd3 | 9537 | -0.045 | 0.0402 | No |
| 77 | Urod | 9693 | -0.050 | 0.0333 | No |
| 78 | Tgfbrap1 | 10069 | -0.066 | 0.0148 | No |
| 79 | Rrad | 10140 | -0.070 | 0.0132 | No |
| 80 | Dnajb1 | 10512 | -0.087 | -0.0044 | No |
| 81 | Bid | 10605 | -0.090 | -0.0066 | No |
| 82 | Btg2 | 10922 | -0.104 | -0.0206 | No |
| 83 | Nxf1 | 10937 | -0.105 | -0.0181 | No |
| 84 | Chka | 10990 | -0.107 | -0.0176 | No |
| 85 | Ppif | 11031 | -0.109 | -0.0164 | No |
| 86 | Plcl1 | 11187 | -0.115 | -0.0212 | No |
| 87 | Ppp1r2 | 11294 | -0.120 | -0.0233 | No |
| 88 | Wiz | 11353 | -0.122 | -0.0226 | No |
| 89 | Ap2s1 | 11366 | -0.123 | -0.0194 | No |
| 90 | Alas1 | 11619 | -0.135 | -0.0289 | No |
| 91 | Mgat1 | 11660 | -0.137 | -0.0269 | No |
| 92 | Sigmar1 | 11732 | -0.140 | -0.0264 | No |
| 93 | Cdkn1c | 11836 | -0.144 | -0.0275 | No |
| 94 | Btg3 | 11843 | -0.145 | -0.0233 | No |
| 95 | Ctsl | 12120 | -0.157 | -0.0334 | No |
| 96 | Selenow | 12277 | -0.165 | -0.0368 | No |
| 97 | Mmp14 | 12292 | -0.165 | -0.0324 | No |
| 98 | Mark2 | 12579 | -0.180 | -0.0424 | No |
| 99 | Icam1 | 12897 | -0.196 | -0.0536 | No |
| 100 | Rhob | 13267 | -0.215 | -0.0670 | No |
| 101 | Tap1 | 13371 | -0.220 | -0.0657 | No |
| 102 | Dgat1 | 13607 | -0.232 | -0.0713 | No |
| 103 | Cnp | 13663 | -0.235 | -0.0669 | No |
| 104 | Slc25a4 | 13681 | -0.236 | -0.0605 | No |
| 105 | Nr4a1 | 13689 | -0.236 | -0.0535 | No |
| 106 | Ppt1 | 13755 | -0.240 | -0.0495 | No |
| 107 | Tst | 13783 | -0.242 | -0.0434 | No |
| 108 | Tuba4a | 13819 | -0.244 | -0.0376 | No |
| 109 | Hspa2 | 14212 | -0.262 | -0.0509 | No |
| 110 | Sod2 | 14215 | -0.263 | -0.0427 | No |
| 111 | H2ax | 14624 | -0.286 | -0.0561 | No |
| 112 | Tmbim6 | 15010 | -0.309 | -0.0674 | No |
| 113 | Hyal2 | 15699 | -0.356 | -0.0939 | No |
| 114 | Cdo1 | 15706 | -0.357 | -0.0830 | No |
| 115 | Eno2 | 15728 | -0.359 | -0.0729 | No |
| 116 | Sqstm1 | 16276 | -0.404 | -0.0901 | No |
| 117 | Tyro3 | 16330 | -0.409 | -0.0802 | No |
| 118 | Slc6a8 | 16505 | -0.425 | -0.0764 | No |
| 119 | Kcnh2 | 16621 | -0.439 | -0.0689 | No |
| 120 | Car2 | 16809 | -0.458 | -0.0648 | No |
| 121 | Nat2 | 16966 | -0.473 | -0.0585 | No |
| 122 | Nfkbia | 17009 | -0.478 | -0.0458 | No |
| 123 | Polg2 | 17194 | -0.500 | -0.0401 | No |
| 124 | Gch1 | 17228 | -0.504 | -0.0261 | No |
| 125 | Ephx1 | 17889 | -0.627 | -0.0426 | No |
| 126 | Grina | 18045 | -0.680 | -0.0297 | No |
| 127 | Furin | 18121 | -0.711 | -0.0115 | No |
| 128 | Rasgrp1 | 18310 | -0.865 | 0.0054 | No |