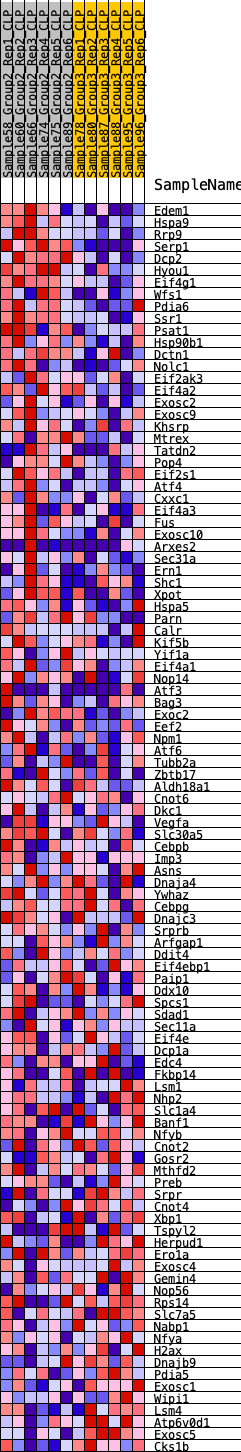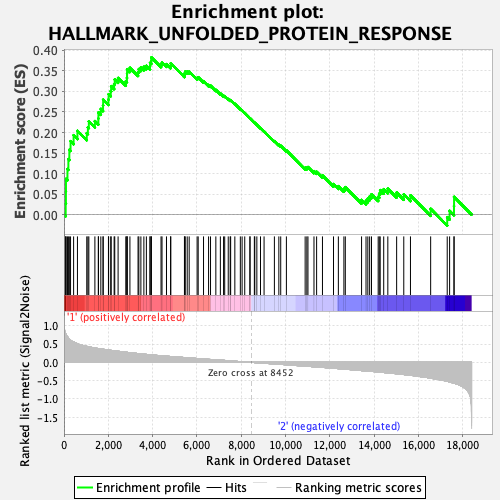
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.38274005 |
| Normalized Enrichment Score (NES) | 1.6058968 |
| Nominal p-value | 0.033932135 |
| FDR q-value | 0.13834642 |
| FWER p-Value | 0.261 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Edem1 | 76 | 0.784 | 0.0269 | Yes |
| 2 | Hspa9 | 86 | 0.769 | 0.0569 | Yes |
| 3 | Rrp9 | 89 | 0.764 | 0.0871 | Yes |
| 4 | Serp1 | 156 | 0.697 | 0.1111 | Yes |
| 5 | Dcp2 | 198 | 0.663 | 0.1352 | Yes |
| 6 | Hyou1 | 241 | 0.632 | 0.1579 | Yes |
| 7 | Eif4g1 | 296 | 0.601 | 0.1788 | Yes |
| 8 | Wfs1 | 434 | 0.555 | 0.1933 | Yes |
| 9 | Pdia6 | 607 | 0.505 | 0.2040 | Yes |
| 10 | Ssr1 | 1028 | 0.435 | 0.1983 | Yes |
| 11 | Psat1 | 1079 | 0.429 | 0.2125 | Yes |
| 12 | Hsp90b1 | 1122 | 0.424 | 0.2270 | Yes |
| 13 | Dctn1 | 1395 | 0.390 | 0.2276 | Yes |
| 14 | Nolc1 | 1551 | 0.375 | 0.2340 | Yes |
| 15 | Eif2ak3 | 1555 | 0.374 | 0.2487 | Yes |
| 16 | Eif4a2 | 1663 | 0.364 | 0.2573 | Yes |
| 17 | Exosc2 | 1763 | 0.357 | 0.2661 | Yes |
| 18 | Exosc9 | 1769 | 0.357 | 0.2799 | Yes |
| 19 | Khsrp | 2007 | 0.334 | 0.2802 | Yes |
| 20 | Mtrex | 2015 | 0.333 | 0.2931 | Yes |
| 21 | Tatdn2 | 2111 | 0.327 | 0.3008 | Yes |
| 22 | Pop4 | 2130 | 0.325 | 0.3127 | Yes |
| 23 | Eif2s1 | 2264 | 0.316 | 0.3180 | Yes |
| 24 | Atf4 | 2292 | 0.314 | 0.3290 | Yes |
| 25 | Cxxc1 | 2443 | 0.301 | 0.3327 | Yes |
| 26 | Eif4a3 | 2791 | 0.276 | 0.3247 | Yes |
| 27 | Fus | 2841 | 0.272 | 0.3328 | Yes |
| 28 | Exosc10 | 2847 | 0.272 | 0.3433 | Yes |
| 29 | Arxes2 | 2850 | 0.272 | 0.3540 | Yes |
| 30 | Sec31a | 2977 | 0.262 | 0.3574 | Yes |
| 31 | Ern1 | 3344 | 0.238 | 0.3469 | Yes |
| 32 | Shc1 | 3380 | 0.235 | 0.3543 | Yes |
| 33 | Xpot | 3472 | 0.230 | 0.3584 | Yes |
| 34 | Hspa5 | 3607 | 0.222 | 0.3599 | Yes |
| 35 | Parn | 3720 | 0.215 | 0.3623 | Yes |
| 36 | Calr | 3884 | 0.207 | 0.3616 | Yes |
| 37 | Kif5b | 3888 | 0.207 | 0.3696 | Yes |
| 38 | Yif1a | 3942 | 0.203 | 0.3748 | Yes |
| 39 | Eif4a1 | 3945 | 0.203 | 0.3827 | Yes |
| 40 | Nop14 | 4382 | 0.179 | 0.3660 | No |
| 41 | Atf3 | 4426 | 0.177 | 0.3707 | No |
| 42 | Bag3 | 4628 | 0.167 | 0.3663 | No |
| 43 | Exoc2 | 4814 | 0.159 | 0.3625 | No |
| 44 | Eef2 | 4822 | 0.159 | 0.3684 | No |
| 45 | Npm1 | 5439 | 0.130 | 0.3399 | No |
| 46 | Atf6 | 5452 | 0.129 | 0.3444 | No |
| 47 | Tubb2a | 5474 | 0.129 | 0.3484 | No |
| 48 | Zbtb17 | 5564 | 0.125 | 0.3484 | No |
| 49 | Aldh18a1 | 5653 | 0.120 | 0.3484 | No |
| 50 | Cnot6 | 6011 | 0.104 | 0.3330 | No |
| 51 | Dkc1 | 6060 | 0.101 | 0.3344 | No |
| 52 | Vegfa | 6301 | 0.091 | 0.3249 | No |
| 53 | Slc30a5 | 6529 | 0.081 | 0.3157 | No |
| 54 | Cebpb | 6617 | 0.077 | 0.3140 | No |
| 55 | Imp3 | 6857 | 0.068 | 0.3036 | No |
| 56 | Asns | 7061 | 0.060 | 0.2949 | No |
| 57 | Dnaja4 | 7206 | 0.054 | 0.2892 | No |
| 58 | Ywhaz | 7257 | 0.052 | 0.2885 | No |
| 59 | Cebpg | 7417 | 0.043 | 0.2815 | No |
| 60 | Dnajc3 | 7504 | 0.040 | 0.2784 | No |
| 61 | Srprb | 7526 | 0.039 | 0.2788 | No |
| 62 | Arfgap1 | 7715 | 0.030 | 0.2697 | No |
| 63 | Ddit4 | 7963 | 0.021 | 0.2571 | No |
| 64 | Eif4ebp1 | 8043 | 0.017 | 0.2534 | No |
| 65 | Paip1 | 8162 | 0.011 | 0.2474 | No |
| 66 | Ddx10 | 8398 | 0.002 | 0.2346 | No |
| 67 | Spcs1 | 8399 | 0.002 | 0.2347 | No |
| 68 | Sdad1 | 8596 | -0.005 | 0.2242 | No |
| 69 | Sec11a | 8616 | -0.006 | 0.2234 | No |
| 70 | Eif4e | 8709 | -0.010 | 0.2188 | No |
| 71 | Dcp1a | 8874 | -0.017 | 0.2105 | No |
| 72 | Edc4 | 9035 | -0.023 | 0.2027 | No |
| 73 | Fkbp14 | 9501 | -0.043 | 0.1790 | No |
| 74 | Lsm1 | 9702 | -0.051 | 0.1701 | No |
| 75 | Nhp2 | 9787 | -0.055 | 0.1677 | No |
| 76 | Slc1a4 | 10043 | -0.065 | 0.1563 | No |
| 77 | Banf1 | 10890 | -0.103 | 0.1142 | No |
| 78 | Nfyb | 10966 | -0.106 | 0.1143 | No |
| 79 | Cnot2 | 11017 | -0.108 | 0.1158 | No |
| 80 | Gosr2 | 11289 | -0.119 | 0.1057 | No |
| 81 | Mthfd2 | 11408 | -0.125 | 0.1042 | No |
| 82 | Preb | 11672 | -0.137 | 0.0953 | No |
| 83 | Srpr | 12171 | -0.159 | 0.0744 | No |
| 84 | Cnot4 | 12390 | -0.171 | 0.0693 | No |
| 85 | Xbp1 | 12640 | -0.183 | 0.0629 | No |
| 86 | Tspyl2 | 12704 | -0.186 | 0.0669 | No |
| 87 | Herpud1 | 13438 | -0.224 | 0.0357 | No |
| 88 | Ero1a | 13639 | -0.234 | 0.0341 | No |
| 89 | Exosc4 | 13721 | -0.238 | 0.0391 | No |
| 90 | Gemin4 | 13803 | -0.243 | 0.0443 | No |
| 91 | Nop56 | 13886 | -0.246 | 0.0496 | No |
| 92 | Rps14 | 14188 | -0.261 | 0.0434 | No |
| 93 | Slc7a5 | 14237 | -0.264 | 0.0513 | No |
| 94 | Nabp1 | 14279 | -0.266 | 0.0596 | No |
| 95 | Nfya | 14438 | -0.276 | 0.0619 | No |
| 96 | H2ax | 14624 | -0.286 | 0.0632 | No |
| 97 | Dnajb9 | 15022 | -0.310 | 0.0538 | No |
| 98 | Pdia5 | 15344 | -0.331 | 0.0494 | No |
| 99 | Exosc1 | 15646 | -0.351 | 0.0468 | No |
| 100 | Wipi1 | 16558 | -0.431 | 0.0142 | No |
| 101 | Lsm4 | 17306 | -0.515 | -0.0062 | No |
| 102 | Atp6v0d1 | 17414 | -0.536 | 0.0092 | No |
| 103 | Exosc5 | 17612 | -0.567 | 0.0209 | No |
| 104 | Cks1b | 17616 | -0.569 | 0.0433 | No |