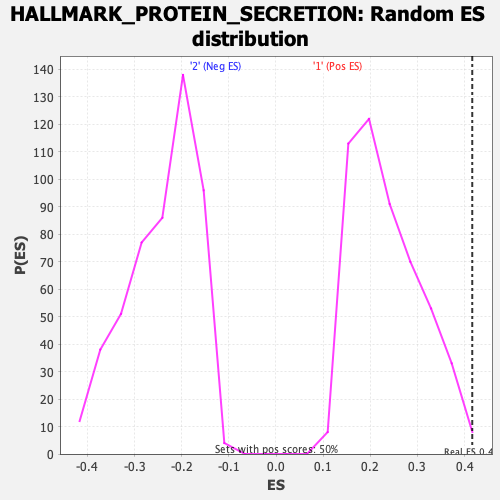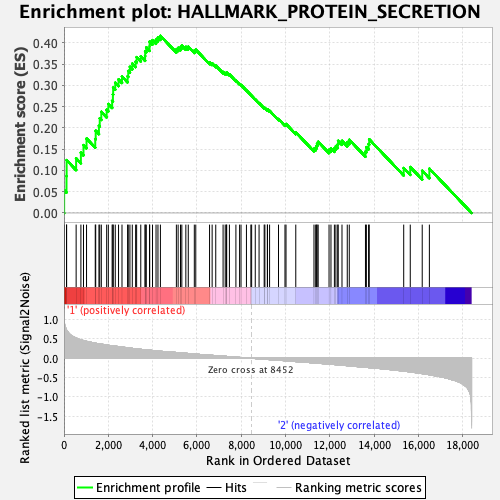
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.41638786 |
| Normalized Enrichment Score (NES) | 1.7744485 |
| Nominal p-value | 0.008032128 |
| FDR q-value | 0.07408978 |
| FWER p-Value | 0.06 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Tspan8 | 7 | 1.027 | 0.0537 | Yes |
| 2 | Abca1 | 113 | 0.733 | 0.0865 | Yes |
| 3 | Cltc | 119 | 0.721 | 0.1242 | Yes |
| 4 | Copb2 | 548 | 0.523 | 0.1284 | Yes |
| 5 | Anp32e | 763 | 0.475 | 0.1417 | Yes |
| 6 | Napg | 880 | 0.457 | 0.1595 | Yes |
| 7 | Atp1a1 | 1018 | 0.437 | 0.1750 | Yes |
| 8 | Egfr | 1411 | 0.388 | 0.1740 | Yes |
| 9 | M6pr | 1432 | 0.386 | 0.1933 | Yes |
| 10 | Uso1 | 1578 | 0.373 | 0.2050 | Yes |
| 11 | Tpd52 | 1614 | 0.370 | 0.2225 | Yes |
| 12 | Zw10 | 1685 | 0.362 | 0.2378 | Yes |
| 13 | Atp7a | 1927 | 0.342 | 0.2426 | Yes |
| 14 | Clta | 2002 | 0.334 | 0.2562 | Yes |
| 15 | Dst | 2174 | 0.322 | 0.2638 | Yes |
| 16 | Copb1 | 2209 | 0.320 | 0.2788 | Yes |
| 17 | Galc | 2221 | 0.318 | 0.2950 | Yes |
| 18 | Tmed10 | 2315 | 0.313 | 0.3063 | Yes |
| 19 | Atp6v1h | 2463 | 0.300 | 0.3141 | Yes |
| 20 | Ocrl | 2619 | 0.288 | 0.3208 | Yes |
| 21 | Vamp3 | 2872 | 0.270 | 0.3213 | Yes |
| 22 | Sec24d | 2901 | 0.268 | 0.3339 | Yes |
| 23 | Sec31a | 2977 | 0.262 | 0.3436 | Yes |
| 24 | Tmx1 | 3084 | 0.255 | 0.3512 | Yes |
| 25 | Rab5a | 3235 | 0.245 | 0.3559 | Yes |
| 26 | Rer1 | 3280 | 0.243 | 0.3663 | Yes |
| 27 | Gla | 3467 | 0.230 | 0.3683 | Yes |
| 28 | Rps6ka3 | 3662 | 0.219 | 0.3692 | Yes |
| 29 | Arfgef2 | 3667 | 0.218 | 0.3804 | Yes |
| 30 | Vps45 | 3716 | 0.215 | 0.3892 | Yes |
| 31 | Stx16 | 3865 | 0.208 | 0.3920 | Yes |
| 32 | Mon2 | 3867 | 0.208 | 0.4029 | Yes |
| 33 | Lman1 | 3997 | 0.199 | 0.4063 | Yes |
| 34 | Arcn1 | 4156 | 0.191 | 0.4078 | Yes |
| 35 | Arf1 | 4243 | 0.186 | 0.4129 | Yes |
| 36 | Cog2 | 4354 | 0.180 | 0.4164 | Yes |
| 37 | Adam10 | 5075 | 0.148 | 0.3849 | No |
| 38 | Cope | 5154 | 0.144 | 0.3882 | No |
| 39 | Ykt6 | 5261 | 0.139 | 0.3897 | No |
| 40 | Arfgef1 | 5321 | 0.135 | 0.3936 | No |
| 41 | Ap2b1 | 5502 | 0.127 | 0.3905 | No |
| 42 | Rab22a | 5612 | 0.123 | 0.3910 | No |
| 43 | Bet1 | 5885 | 0.109 | 0.3819 | No |
| 44 | Golga4 | 5951 | 0.106 | 0.3839 | No |
| 45 | Krt18 | 6573 | 0.079 | 0.3542 | No |
| 46 | Lamp2 | 6690 | 0.075 | 0.3518 | No |
| 47 | Rab2a | 6853 | 0.068 | 0.3465 | No |
| 48 | Mapk1 | 7184 | 0.055 | 0.3314 | No |
| 49 | Snap23 | 7280 | 0.050 | 0.3289 | No |
| 50 | Bnip3 | 7332 | 0.048 | 0.3286 | No |
| 51 | Pam | 7347 | 0.046 | 0.3303 | No |
| 52 | Stx7 | 7470 | 0.041 | 0.3258 | No |
| 53 | Ctsc | 7763 | 0.028 | 0.3113 | No |
| 54 | Ap1g1 | 7930 | 0.022 | 0.3034 | No |
| 55 | Scamp1 | 7994 | 0.019 | 0.3010 | No |
| 56 | Scamp3 | 8239 | 0.009 | 0.2881 | No |
| 57 | Ergic3 | 8439 | 0.000 | 0.2773 | No |
| 58 | Clcn3 | 8474 | -0.000 | 0.2754 | No |
| 59 | Rab9 | 8637 | -0.007 | 0.2670 | No |
| 60 | Ap3b1 | 8809 | -0.014 | 0.2584 | No |
| 61 | Snx2 | 9040 | -0.024 | 0.2470 | No |
| 62 | Igf2r | 9072 | -0.025 | 0.2467 | No |
| 63 | Ap2m1 | 9183 | -0.030 | 0.2422 | No |
| 64 | Napa | 9190 | -0.030 | 0.2435 | No |
| 65 | Arfip1 | 9284 | -0.034 | 0.2402 | No |
| 66 | Dnm1l | 9687 | -0.050 | 0.2209 | No |
| 67 | Tsg101 | 9979 | -0.063 | 0.2083 | No |
| 68 | Tom1l1 | 10026 | -0.064 | 0.2092 | No |
| 69 | Yipf6 | 10467 | -0.085 | 0.1896 | No |
| 70 | Gosr2 | 11289 | -0.119 | 0.1511 | No |
| 71 | Ap2s1 | 11366 | -0.123 | 0.1534 | No |
| 72 | Sec22b | 11407 | -0.125 | 0.1578 | No |
| 73 | Dop1a | 11431 | -0.126 | 0.1632 | No |
| 74 | Vps4b | 11478 | -0.128 | 0.1674 | No |
| 75 | Rab14 | 11969 | -0.151 | 0.1486 | No |
| 76 | Cav2 | 12056 | -0.154 | 0.1520 | No |
| 77 | Sgms1 | 12211 | -0.162 | 0.1521 | No |
| 78 | Ap3s1 | 12279 | -0.165 | 0.1572 | No |
| 79 | Stam | 12364 | -0.170 | 0.1615 | No |
| 80 | Vamp4 | 12380 | -0.170 | 0.1696 | No |
| 81 | Gnas | 12554 | -0.179 | 0.1696 | No |
| 82 | Gbf1 | 12792 | -0.191 | 0.1667 | No |
| 83 | Stx12 | 12883 | -0.195 | 0.1721 | No |
| 84 | Cln5 | 13614 | -0.233 | 0.1445 | No |
| 85 | Cd63 | 13655 | -0.235 | 0.1546 | No |
| 86 | Ppt1 | 13755 | -0.240 | 0.1619 | No |
| 87 | Arfgap3 | 13784 | -0.242 | 0.1731 | No |
| 88 | Kif1b | 15341 | -0.331 | 0.1056 | No |
| 89 | Sod1 | 15637 | -0.351 | 0.1079 | No |
| 90 | Sh3gl2 | 16177 | -0.396 | 0.0994 | No |
| 91 | Ica1 | 16500 | -0.425 | 0.1042 | No |