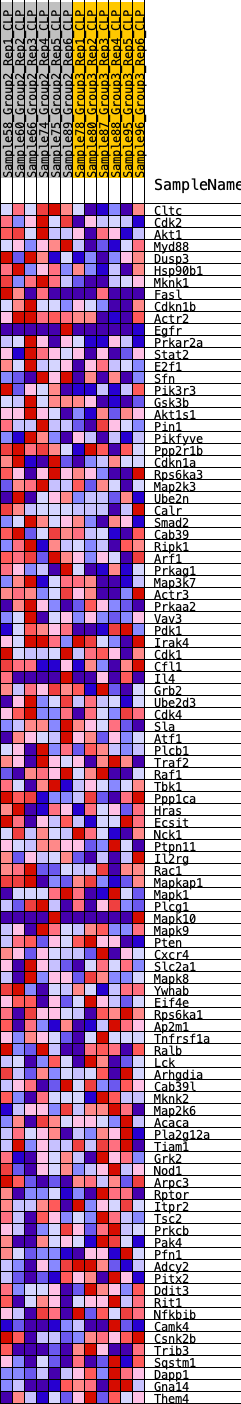
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
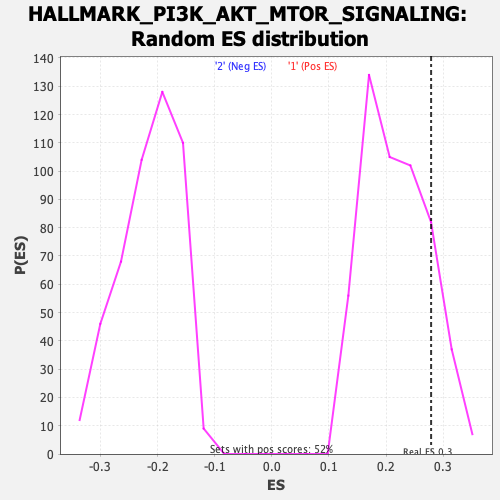
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.27868092 |
| Normalized Enrichment Score (NES) | 1.2805537 |
| Nominal p-value | 0.15869981 |
| FDR q-value | 0.57441366 |
| FWER p-Value | 0.834 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cltc | 119 | 0.721 | 0.0293 | Yes |
| 2 | Cdk2 | 642 | 0.497 | 0.0255 | Yes |
| 3 | Akt1 | 740 | 0.478 | 0.0439 | Yes |
| 4 | Myd88 | 962 | 0.444 | 0.0539 | Yes |
| 5 | Dusp3 | 995 | 0.439 | 0.0740 | Yes |
| 6 | Hsp90b1 | 1122 | 0.424 | 0.0882 | Yes |
| 7 | Mknk1 | 1133 | 0.422 | 0.1086 | Yes |
| 8 | Fasl | 1183 | 0.415 | 0.1265 | Yes |
| 9 | Cdkn1b | 1281 | 0.403 | 0.1412 | Yes |
| 10 | Actr2 | 1385 | 0.391 | 0.1550 | Yes |
| 11 | Egfr | 1411 | 0.388 | 0.1729 | Yes |
| 12 | Prkar2a | 1516 | 0.378 | 0.1860 | Yes |
| 13 | Stat2 | 1569 | 0.374 | 0.2018 | Yes |
| 14 | E2f1 | 1618 | 0.369 | 0.2175 | Yes |
| 15 | Sfn | 1721 | 0.360 | 0.2298 | Yes |
| 16 | Pik3r3 | 2779 | 0.277 | 0.1858 | Yes |
| 17 | Gsk3b | 2896 | 0.268 | 0.1928 | Yes |
| 18 | Akt1s1 | 3246 | 0.245 | 0.1859 | Yes |
| 19 | Pin1 | 3345 | 0.238 | 0.1923 | Yes |
| 20 | Pikfyve | 3396 | 0.235 | 0.2012 | Yes |
| 21 | Ppp2r1b | 3497 | 0.228 | 0.2071 | Yes |
| 22 | Cdkn1a | 3579 | 0.224 | 0.2138 | Yes |
| 23 | Rps6ka3 | 3662 | 0.219 | 0.2201 | Yes |
| 24 | Map2k3 | 3882 | 0.207 | 0.2184 | Yes |
| 25 | Ube2n | 3883 | 0.207 | 0.2287 | Yes |
| 26 | Calr | 3884 | 0.207 | 0.2390 | Yes |
| 27 | Smad2 | 3913 | 0.205 | 0.2477 | Yes |
| 28 | Cab39 | 4035 | 0.198 | 0.2509 | Yes |
| 29 | Ripk1 | 4188 | 0.189 | 0.2520 | Yes |
| 30 | Arf1 | 4243 | 0.186 | 0.2582 | Yes |
| 31 | Prkag1 | 4300 | 0.184 | 0.2643 | Yes |
| 32 | Map3k7 | 4476 | 0.174 | 0.2634 | Yes |
| 33 | Actr3 | 4556 | 0.171 | 0.2676 | Yes |
| 34 | Prkaa2 | 4580 | 0.169 | 0.2747 | Yes |
| 35 | Vav3 | 4868 | 0.156 | 0.2668 | Yes |
| 36 | Pdk1 | 5036 | 0.150 | 0.2651 | Yes |
| 37 | Irak4 | 5049 | 0.149 | 0.2719 | Yes |
| 38 | Cdk1 | 5084 | 0.147 | 0.2773 | Yes |
| 39 | Cfl1 | 5206 | 0.142 | 0.2778 | Yes |
| 40 | Il4 | 5314 | 0.136 | 0.2787 | Yes |
| 41 | Grb2 | 5490 | 0.128 | 0.2755 | No |
| 42 | Ube2d3 | 5549 | 0.125 | 0.2785 | No |
| 43 | Cdk4 | 5661 | 0.120 | 0.2784 | No |
| 44 | Sla | 5780 | 0.114 | 0.2777 | No |
| 45 | Atf1 | 6006 | 0.104 | 0.2705 | No |
| 46 | Plcb1 | 6390 | 0.087 | 0.2540 | No |
| 47 | Traf2 | 6539 | 0.080 | 0.2499 | No |
| 48 | Raf1 | 6699 | 0.074 | 0.2449 | No |
| 49 | Tbk1 | 6744 | 0.072 | 0.2461 | No |
| 50 | Ppp1ca | 6771 | 0.071 | 0.2482 | No |
| 51 | Hras | 6833 | 0.069 | 0.2483 | No |
| 52 | Ecsit | 6975 | 0.064 | 0.2437 | No |
| 53 | Nck1 | 6991 | 0.063 | 0.2461 | No |
| 54 | Ptpn11 | 6993 | 0.063 | 0.2491 | No |
| 55 | Il2rg | 7081 | 0.058 | 0.2473 | No |
| 56 | Rac1 | 7118 | 0.057 | 0.2481 | No |
| 57 | Mapkap1 | 7139 | 0.056 | 0.2498 | No |
| 58 | Mapk1 | 7184 | 0.055 | 0.2501 | No |
| 59 | Plcg1 | 7495 | 0.040 | 0.2352 | No |
| 60 | Mapk10 | 7903 | 0.022 | 0.2141 | No |
| 61 | Mapk9 | 8153 | 0.012 | 0.2011 | No |
| 62 | Pten | 8166 | 0.011 | 0.2010 | No |
| 63 | Cxcr4 | 8377 | 0.003 | 0.1896 | No |
| 64 | Slc2a1 | 8567 | -0.004 | 0.1795 | No |
| 65 | Mapk8 | 8587 | -0.005 | 0.1787 | No |
| 66 | Ywhab | 8658 | -0.008 | 0.1753 | No |
| 67 | Eif4e | 8709 | -0.010 | 0.1731 | No |
| 68 | Rps6ka1 | 9008 | -0.022 | 0.1579 | No |
| 69 | Ap2m1 | 9183 | -0.030 | 0.1499 | No |
| 70 | Tnfrsf1a | 9359 | -0.038 | 0.1422 | No |
| 71 | Ralb | 9573 | -0.045 | 0.1328 | No |
| 72 | Lck | 9586 | -0.046 | 0.1345 | No |
| 73 | Arhgdia | 9819 | -0.056 | 0.1246 | No |
| 74 | Cab39l | 10149 | -0.070 | 0.1101 | No |
| 75 | Mknk2 | 10911 | -0.104 | 0.0736 | No |
| 76 | Map2k6 | 10916 | -0.104 | 0.0786 | No |
| 77 | Acaca | 11028 | -0.109 | 0.0779 | No |
| 78 | Pla2g12a | 11448 | -0.127 | 0.0613 | No |
| 79 | Tiam1 | 11560 | -0.132 | 0.0618 | No |
| 80 | Grk2 | 11597 | -0.134 | 0.0665 | No |
| 81 | Nod1 | 11782 | -0.142 | 0.0635 | No |
| 82 | Arpc3 | 11946 | -0.149 | 0.0620 | No |
| 83 | Rptor | 12113 | -0.156 | 0.0607 | No |
| 84 | Itpr2 | 12136 | -0.157 | 0.0673 | No |
| 85 | Tsc2 | 12151 | -0.158 | 0.0744 | No |
| 86 | Prkcb | 13348 | -0.219 | 0.0200 | No |
| 87 | Pak4 | 13776 | -0.241 | 0.0086 | No |
| 88 | Pfn1 | 14040 | -0.254 | 0.0069 | No |
| 89 | Adcy2 | 14042 | -0.254 | 0.0195 | No |
| 90 | Pitx2 | 14457 | -0.278 | 0.0107 | No |
| 91 | Ddit3 | 14775 | -0.295 | 0.0080 | No |
| 92 | Rit1 | 15527 | -0.344 | -0.0160 | No |
| 93 | Nfkbib | 16035 | -0.382 | -0.0247 | No |
| 94 | Camk4 | 16085 | -0.388 | -0.0081 | No |
| 95 | Csnk2b | 16102 | -0.390 | 0.0104 | No |
| 96 | Trib3 | 16226 | -0.399 | 0.0235 | No |
| 97 | Sqstm1 | 16276 | -0.404 | 0.0409 | No |
| 98 | Dapp1 | 16850 | -0.463 | 0.0326 | No |
| 99 | Gna14 | 17295 | -0.513 | 0.0338 | No |
| 100 | Them4 | 17439 | -0.542 | 0.0529 | No |