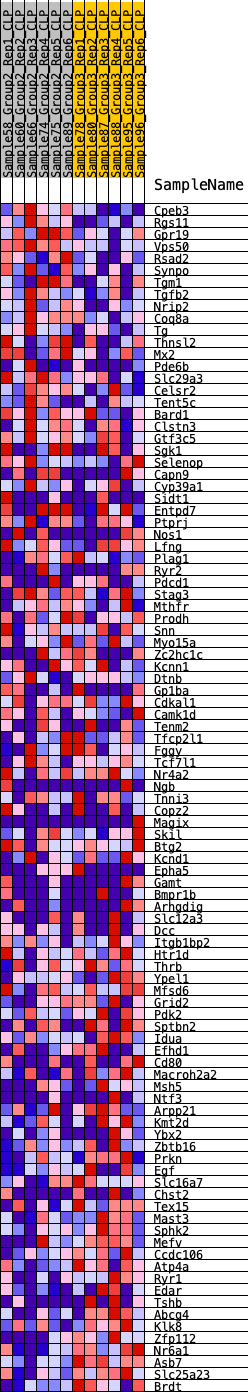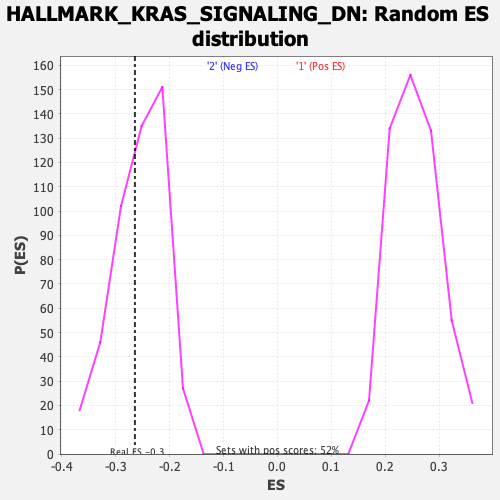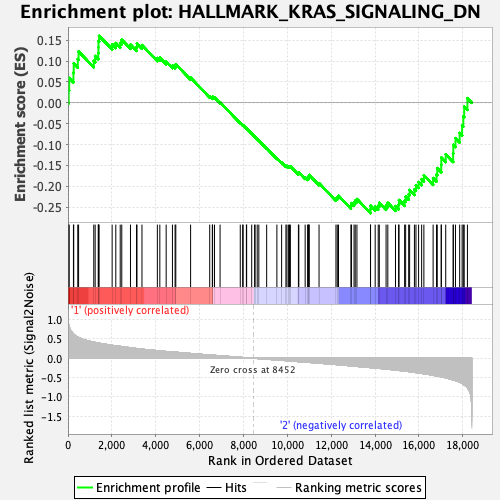
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.26427376 |
| Normalized Enrichment Score (NES) | -1.0354635 |
| Nominal p-value | 0.38204592 |
| FDR q-value | 0.93820906 |
| FWER p-Value | 0.985 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cpeb3 | 31 | 0.871 | 0.0306 | No |
| 2 | Rgs11 | 53 | 0.812 | 0.0595 | No |
| 3 | Gpr19 | 248 | 0.628 | 0.0721 | No |
| 4 | Vps50 | 261 | 0.619 | 0.0944 | No |
| 5 | Rsad2 | 446 | 0.550 | 0.1047 | No |
| 6 | Synpo | 483 | 0.539 | 0.1227 | No |
| 7 | Tgm1 | 1175 | 0.416 | 0.1004 | No |
| 8 | Tgfb2 | 1238 | 0.408 | 0.1121 | No |
| 9 | Nrip2 | 1380 | 0.391 | 0.1189 | No |
| 10 | Coq8a | 1388 | 0.391 | 0.1330 | No |
| 11 | Tg | 1390 | 0.391 | 0.1474 | No |
| 12 | Thnsl2 | 1418 | 0.388 | 0.1603 | No |
| 13 | Mx2 | 2012 | 0.333 | 0.1402 | No |
| 14 | Pde6b | 2179 | 0.322 | 0.1431 | No |
| 15 | Slc29a3 | 2380 | 0.307 | 0.1435 | No |
| 16 | Celsr2 | 2451 | 0.301 | 0.1508 | No |
| 17 | Tent5c | 2848 | 0.272 | 0.1393 | No |
| 18 | Bard1 | 3124 | 0.251 | 0.1335 | No |
| 19 | Clstn3 | 3144 | 0.250 | 0.1418 | No |
| 20 | Gtf3c5 | 3372 | 0.236 | 0.1381 | No |
| 21 | Sgk1 | 4073 | 0.196 | 0.1071 | No |
| 22 | Selenop | 4186 | 0.189 | 0.1080 | No |
| 23 | Capn9 | 4475 | 0.174 | 0.0987 | No |
| 24 | Cyp39a1 | 4761 | 0.161 | 0.0891 | No |
| 25 | Sidt1 | 4881 | 0.156 | 0.0884 | No |
| 26 | Entpd7 | 4914 | 0.155 | 0.0924 | No |
| 27 | Ptprj | 5589 | 0.124 | 0.0601 | No |
| 28 | Nos1 | 6460 | 0.084 | 0.0157 | No |
| 29 | Lfng | 6579 | 0.079 | 0.0122 | No |
| 30 | Plag1 | 6591 | 0.078 | 0.0145 | No |
| 31 | Ryr2 | 6679 | 0.075 | 0.0126 | No |
| 32 | Pdcd1 | 6932 | 0.065 | 0.0012 | No |
| 33 | Stag3 | 7853 | 0.024 | -0.0482 | No |
| 34 | Mthfr | 7964 | 0.021 | -0.0534 | No |
| 35 | Prodh | 7982 | 0.020 | -0.0536 | No |
| 36 | Snn | 8132 | 0.013 | -0.0613 | No |
| 37 | Myo15a | 8144 | 0.012 | -0.0614 | No |
| 38 | Zc2hc1c | 8374 | 0.003 | -0.0738 | No |
| 39 | Kcnn1 | 8509 | -0.002 | -0.0811 | No |
| 40 | Dtnb | 8527 | -0.002 | -0.0819 | No |
| 41 | Gp1ba | 8625 | -0.007 | -0.0870 | No |
| 42 | Cdkal1 | 8699 | -0.010 | -0.0906 | No |
| 43 | Camk1d | 9056 | -0.024 | -0.1091 | No |
| 44 | Tenm2 | 9522 | -0.044 | -0.1329 | No |
| 45 | Tfcp2l1 | 9738 | -0.052 | -0.1427 | No |
| 46 | Fggy | 9934 | -0.060 | -0.1511 | No |
| 47 | Tcf7l1 | 9981 | -0.063 | -0.1513 | No |
| 48 | Nr4a2 | 10056 | -0.066 | -0.1529 | No |
| 49 | Ngb | 10098 | -0.068 | -0.1527 | No |
| 50 | Tnni3 | 10137 | -0.069 | -0.1522 | No |
| 51 | Copz2 | 10503 | -0.086 | -0.1689 | No |
| 52 | Magix | 10522 | -0.087 | -0.1666 | No |
| 53 | Skil | 10811 | -0.099 | -0.1787 | No |
| 54 | Btg2 | 10922 | -0.104 | -0.1809 | No |
| 55 | Kcnd1 | 10968 | -0.106 | -0.1794 | No |
| 56 | Epha5 | 10975 | -0.106 | -0.1758 | No |
| 57 | Gamt | 10996 | -0.107 | -0.1729 | No |
| 58 | Bmpr1b | 11444 | -0.127 | -0.1926 | No |
| 59 | Arhgdig | 12215 | -0.162 | -0.2287 | No |
| 60 | Slc12a3 | 12276 | -0.165 | -0.2259 | No |
| 61 | Dcc | 12331 | -0.168 | -0.2226 | No |
| 62 | Itgb1bp2 | 12896 | -0.196 | -0.2462 | No |
| 63 | Htr1d | 12925 | -0.197 | -0.2404 | No |
| 64 | Thrb | 13040 | -0.205 | -0.2390 | No |
| 65 | Ypel1 | 13093 | -0.208 | -0.2342 | No |
| 66 | Mfsd6 | 13171 | -0.211 | -0.2306 | No |
| 67 | Grid2 | 13789 | -0.242 | -0.2553 | Yes |
| 68 | Pdk2 | 13792 | -0.242 | -0.2465 | Yes |
| 69 | Sptbn2 | 14003 | -0.252 | -0.2486 | Yes |
| 70 | Idua | 14142 | -0.259 | -0.2466 | Yes |
| 71 | Efhd1 | 14191 | -0.261 | -0.2395 | Yes |
| 72 | Cd80 | 14500 | -0.280 | -0.2460 | Yes |
| 73 | Macroh2a2 | 14579 | -0.284 | -0.2397 | Yes |
| 74 | Msh5 | 14929 | -0.305 | -0.2475 | Yes |
| 75 | Ntf3 | 15072 | -0.313 | -0.2437 | Yes |
| 76 | Arpp21 | 15089 | -0.314 | -0.2329 | Yes |
| 77 | Kmt2d | 15345 | -0.331 | -0.2346 | Yes |
| 78 | Ybx2 | 15392 | -0.335 | -0.2247 | Yes |
| 79 | Zbtb16 | 15530 | -0.344 | -0.2194 | Yes |
| 80 | Prkn | 15573 | -0.347 | -0.2089 | Yes |
| 81 | Egf | 15792 | -0.364 | -0.2073 | Yes |
| 82 | Slc16a7 | 15862 | -0.370 | -0.1974 | Yes |
| 83 | Chst2 | 15982 | -0.378 | -0.1899 | Yes |
| 84 | Tex15 | 16121 | -0.391 | -0.1829 | Yes |
| 85 | Mast3 | 16223 | -0.399 | -0.1737 | Yes |
| 86 | Sphk2 | 16644 | -0.441 | -0.1803 | Yes |
| 87 | Mefv | 16799 | -0.457 | -0.1718 | Yes |
| 88 | Ccdc106 | 16832 | -0.461 | -0.1564 | Yes |
| 89 | Atp4a | 17014 | -0.478 | -0.1486 | Yes |
| 90 | Ryr1 | 17018 | -0.478 | -0.1311 | Yes |
| 91 | Edar | 17219 | -0.503 | -0.1234 | Yes |
| 92 | Tshb | 17558 | -0.559 | -0.1211 | Yes |
| 93 | Abcg4 | 17565 | -0.560 | -0.1007 | Yes |
| 94 | Klk8 | 17663 | -0.575 | -0.0847 | Yes |
| 95 | Zfp112 | 17855 | -0.618 | -0.0723 | Yes |
| 96 | Nr6a1 | 17966 | -0.650 | -0.0542 | Yes |
| 97 | Asb7 | 18020 | -0.669 | -0.0323 | Yes |
| 98 | Slc25a23 | 18061 | -0.688 | -0.0091 | Yes |
| 99 | Brdt | 18208 | -0.755 | 0.0109 | Yes |