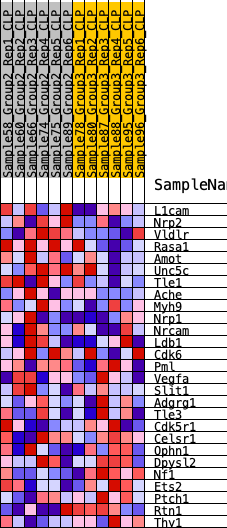
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
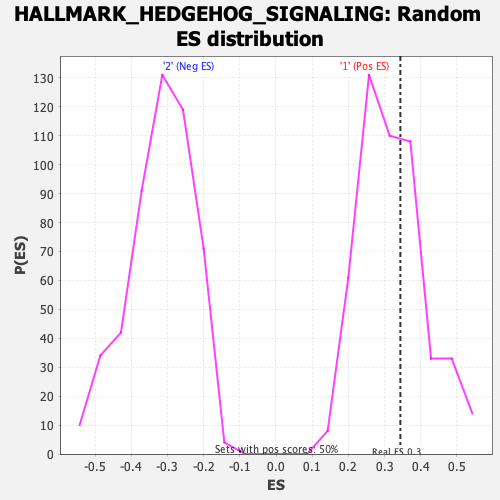
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.34362626 |
| Normalized Enrichment Score (NES) | 1.0738314 |
| Nominal p-value | 0.3694779 |
| FDR q-value | 0.86962765 |
| FWER p-Value | 0.98 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | L1cam | 829 | 0.465 | 0.0280 | Yes |
| 2 | Nrp2 | 853 | 0.460 | 0.0991 | Yes |
| 3 | Vldlr | 1296 | 0.401 | 0.1381 | Yes |
| 4 | Rasa1 | 1535 | 0.377 | 0.1844 | Yes |
| 5 | Amot | 1542 | 0.376 | 0.2431 | Yes |
| 6 | Unc5c | 1632 | 0.368 | 0.2961 | Yes |
| 7 | Tle1 | 1875 | 0.346 | 0.3374 | Yes |
| 8 | Ache | 2599 | 0.290 | 0.3436 | Yes |
| 9 | Myh9 | 3707 | 0.216 | 0.3173 | No |
| 10 | Nrp1 | 4251 | 0.186 | 0.3170 | No |
| 11 | Nrcam | 5204 | 0.142 | 0.2875 | No |
| 12 | Ldb1 | 5535 | 0.126 | 0.2894 | No |
| 13 | Cdk6 | 5994 | 0.105 | 0.2809 | No |
| 14 | Pml | 6155 | 0.097 | 0.2874 | No |
| 15 | Vegfa | 6301 | 0.091 | 0.2938 | No |
| 16 | Slit1 | 7015 | 0.062 | 0.2648 | No |
| 17 | Adgrg1 | 8867 | -0.016 | 0.1667 | No |
| 18 | Tle3 | 9304 | -0.035 | 0.1485 | No |
| 19 | Cdk5r1 | 9970 | -0.062 | 0.1221 | No |
| 20 | Celsr1 | 11133 | -0.113 | 0.0767 | No |
| 21 | Ophn1 | 11287 | -0.119 | 0.0871 | No |
| 22 | Dpysl2 | 11965 | -0.151 | 0.0739 | No |
| 23 | Nf1 | 13310 | -0.217 | 0.0349 | No |
| 24 | Ets2 | 14532 | -0.281 | 0.0127 | No |
| 25 | Ptch1 | 15906 | -0.372 | -0.0034 | No |
| 26 | Rtn1 | 16275 | -0.404 | 0.0401 | No |
| 27 | Thy1 | 17052 | -0.483 | 0.0738 | No |