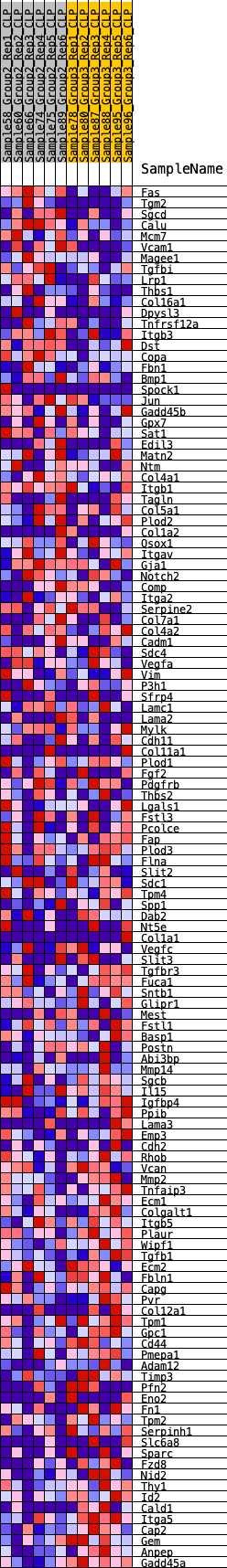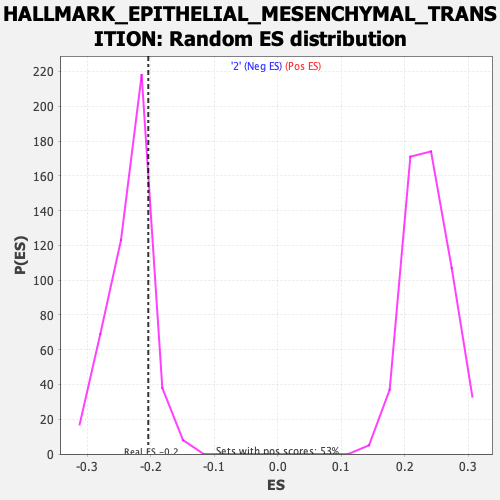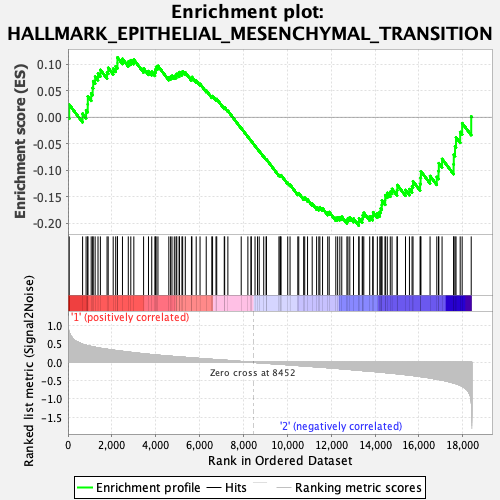
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | -0.20403811 |
| Normalized Enrichment Score (NES) | -0.87647325 |
| Nominal p-value | 0.8308668 |
| FDR q-value | 0.7803571 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fas | 55 | 0.810 | 0.0237 | No |
| 2 | Tgm2 | 665 | 0.492 | 0.0066 | No |
| 3 | Sgcd | 825 | 0.466 | 0.0132 | No |
| 4 | Calu | 900 | 0.455 | 0.0242 | No |
| 5 | Mcm7 | 904 | 0.455 | 0.0390 | No |
| 6 | Vcam1 | 1057 | 0.432 | 0.0449 | No |
| 7 | Magee1 | 1124 | 0.423 | 0.0553 | No |
| 8 | Tgfbi | 1148 | 0.420 | 0.0679 | No |
| 9 | Lrp1 | 1237 | 0.409 | 0.0765 | No |
| 10 | Thbs1 | 1368 | 0.392 | 0.0823 | No |
| 11 | Col16a1 | 1476 | 0.381 | 0.0891 | No |
| 12 | Dpysl3 | 1780 | 0.356 | 0.0842 | No |
| 13 | Tnfrsf12a | 1832 | 0.350 | 0.0930 | No |
| 14 | Itgb3 | 2060 | 0.330 | 0.0915 | No |
| 15 | Dst | 2174 | 0.322 | 0.0959 | No |
| 16 | Copa | 2253 | 0.317 | 0.1021 | No |
| 17 | Fbn1 | 2254 | 0.317 | 0.1125 | No |
| 18 | Bmp1 | 2483 | 0.299 | 0.1099 | No |
| 19 | Spock1 | 2748 | 0.279 | 0.1046 | No |
| 20 | Jun | 2863 | 0.271 | 0.1073 | No |
| 21 | Gadd45b | 3001 | 0.260 | 0.1084 | No |
| 22 | Gpx7 | 3446 | 0.232 | 0.0918 | No |
| 23 | Sat1 | 3669 | 0.218 | 0.0868 | No |
| 24 | Edil3 | 3817 | 0.210 | 0.0857 | No |
| 25 | Matn2 | 3964 | 0.202 | 0.0844 | No |
| 26 | Ntm | 3984 | 0.200 | 0.0899 | No |
| 27 | Col4a1 | 4017 | 0.198 | 0.0947 | No |
| 28 | Itgb1 | 4100 | 0.194 | 0.0966 | No |
| 29 | Tagln | 4596 | 0.169 | 0.0751 | No |
| 30 | Col5a1 | 4677 | 0.165 | 0.0762 | No |
| 31 | Plod2 | 4740 | 0.162 | 0.0781 | No |
| 32 | Col1a2 | 4846 | 0.157 | 0.0775 | No |
| 33 | Qsox1 | 4926 | 0.155 | 0.0783 | No |
| 34 | Itgav | 4959 | 0.153 | 0.0816 | No |
| 35 | Gja1 | 5066 | 0.148 | 0.0807 | No |
| 36 | Notch2 | 5080 | 0.148 | 0.0849 | No |
| 37 | Comp | 5195 | 0.142 | 0.0833 | No |
| 38 | Itga2 | 5226 | 0.140 | 0.0863 | No |
| 39 | Serpine2 | 5347 | 0.134 | 0.0841 | No |
| 40 | Col7a1 | 5628 | 0.122 | 0.0728 | No |
| 41 | Col4a2 | 5649 | 0.121 | 0.0757 | No |
| 42 | Cadm1 | 5844 | 0.111 | 0.0688 | No |
| 43 | Sdc4 | 6019 | 0.103 | 0.0627 | No |
| 44 | Vegfa | 6301 | 0.091 | 0.0503 | No |
| 45 | Vim | 6556 | 0.080 | 0.0390 | No |
| 46 | P3h1 | 6590 | 0.078 | 0.0398 | No |
| 47 | Sfrp4 | 6747 | 0.072 | 0.0336 | No |
| 48 | Lamc1 | 6780 | 0.071 | 0.0342 | No |
| 49 | Lama2 | 7123 | 0.057 | 0.0174 | No |
| 50 | Mylk | 7142 | 0.056 | 0.0183 | No |
| 51 | Cdh11 | 7288 | 0.050 | 0.0120 | No |
| 52 | Col11a1 | 7896 | 0.022 | -0.0205 | No |
| 53 | Plod1 | 8200 | 0.010 | -0.0367 | No |
| 54 | Fgf2 | 8340 | 0.004 | -0.0442 | No |
| 55 | Pdgfrb | 8356 | 0.004 | -0.0449 | No |
| 56 | Thbs2 | 8526 | -0.002 | -0.0540 | No |
| 57 | Lgals1 | 8639 | -0.007 | -0.0599 | No |
| 58 | Fstl3 | 8716 | -0.011 | -0.0637 | No |
| 59 | Pcolce | 8922 | -0.019 | -0.0743 | No |
| 60 | Fap | 9031 | -0.023 | -0.0795 | No |
| 61 | Plod3 | 9047 | -0.024 | -0.0795 | No |
| 62 | Flna | 9616 | -0.047 | -0.1090 | No |
| 63 | Slit2 | 9664 | -0.049 | -0.1100 | No |
| 64 | Sdc1 | 9686 | -0.050 | -0.1095 | No |
| 65 | Tpm4 | 9712 | -0.051 | -0.1092 | No |
| 66 | Spp1 | 10016 | -0.064 | -0.1236 | No |
| 67 | Dab2 | 10122 | -0.069 | -0.1271 | No |
| 68 | Nt5e | 10476 | -0.085 | -0.1436 | No |
| 69 | Col1a1 | 10519 | -0.087 | -0.1430 | No |
| 70 | Vegfc | 10743 | -0.096 | -0.1521 | No |
| 71 | Slit3 | 10791 | -0.098 | -0.1514 | No |
| 72 | Tgfbr3 | 10920 | -0.104 | -0.1550 | No |
| 73 | Fuca1 | 11132 | -0.113 | -0.1628 | No |
| 74 | Sntb1 | 11333 | -0.121 | -0.1697 | No |
| 75 | Glipr1 | 11435 | -0.126 | -0.1711 | No |
| 76 | Mest | 11482 | -0.129 | -0.1694 | No |
| 77 | Fstl1 | 11604 | -0.134 | -0.1716 | No |
| 78 | Basp1 | 11839 | -0.144 | -0.1796 | No |
| 79 | Postn | 11907 | -0.148 | -0.1784 | No |
| 80 | Abi3bp | 12208 | -0.162 | -0.1895 | No |
| 81 | Mmp14 | 12292 | -0.165 | -0.1886 | No |
| 82 | Sgcb | 12392 | -0.171 | -0.1884 | No |
| 83 | Il15 | 12480 | -0.175 | -0.1873 | No |
| 84 | Igfbp4 | 12715 | -0.186 | -0.1940 | No |
| 85 | Ppib | 12762 | -0.189 | -0.1903 | No |
| 86 | Lama3 | 12846 | -0.194 | -0.1884 | No |
| 87 | Emp3 | 13015 | -0.203 | -0.1909 | No |
| 88 | Cdh2 | 13256 | -0.215 | -0.1970 | Yes |
| 89 | Rhob | 13267 | -0.215 | -0.1904 | Yes |
| 90 | Vcan | 13413 | -0.223 | -0.1910 | Yes |
| 91 | Mmp2 | 13435 | -0.224 | -0.1847 | Yes |
| 92 | Tnfaip3 | 13484 | -0.227 | -0.1799 | Yes |
| 93 | Ecm1 | 13753 | -0.240 | -0.1866 | Yes |
| 94 | Colgalt1 | 13890 | -0.246 | -0.1860 | Yes |
| 95 | Itgb5 | 13910 | -0.248 | -0.1788 | Yes |
| 96 | Plaur | 14102 | -0.257 | -0.1808 | Yes |
| 97 | Wipf1 | 14214 | -0.263 | -0.1782 | Yes |
| 98 | Tgfb1 | 14256 | -0.265 | -0.1717 | Yes |
| 99 | Ecm2 | 14304 | -0.268 | -0.1655 | Yes |
| 100 | Fbln1 | 14310 | -0.268 | -0.1569 | Yes |
| 101 | Capg | 14459 | -0.278 | -0.1558 | Yes |
| 102 | Pvr | 14461 | -0.278 | -0.1467 | Yes |
| 103 | Col12a1 | 14554 | -0.283 | -0.1424 | Yes |
| 104 | Tpm1 | 14691 | -0.290 | -0.1403 | Yes |
| 105 | Gpc1 | 14771 | -0.295 | -0.1349 | Yes |
| 106 | Cd44 | 15000 | -0.309 | -0.1372 | Yes |
| 107 | Pmepa1 | 15012 | -0.310 | -0.1276 | Yes |
| 108 | Adam12 | 15385 | -0.334 | -0.1369 | Yes |
| 109 | Timp3 | 15565 | -0.346 | -0.1353 | Yes |
| 110 | Pfn2 | 15684 | -0.355 | -0.1301 | Yes |
| 111 | Eno2 | 15728 | -0.359 | -0.1206 | Yes |
| 112 | Fn1 | 16050 | -0.383 | -0.1255 | Yes |
| 113 | Tpm2 | 16072 | -0.387 | -0.1139 | Yes |
| 114 | Serpinh1 | 16087 | -0.388 | -0.1019 | Yes |
| 115 | Slc6a8 | 16505 | -0.425 | -0.1107 | Yes |
| 116 | Sparc | 16812 | -0.458 | -0.1123 | Yes |
| 117 | Fzd8 | 16890 | -0.467 | -0.1011 | Yes |
| 118 | Nid2 | 16905 | -0.468 | -0.0865 | Yes |
| 119 | Thy1 | 17052 | -0.483 | -0.0785 | Yes |
| 120 | Id2 | 17578 | -0.562 | -0.0887 | Yes |
| 121 | Cald1 | 17587 | -0.563 | -0.0706 | Yes |
| 122 | Itga5 | 17643 | -0.571 | -0.0548 | Yes |
| 123 | Cap2 | 17688 | -0.580 | -0.0381 | Yes |
| 124 | Gem | 17878 | -0.624 | -0.0278 | Yes |
| 125 | Anpep | 17968 | -0.651 | -0.0112 | Yes |
| 126 | Gadd45a | 18381 | -1.070 | 0.0015 | Yes |