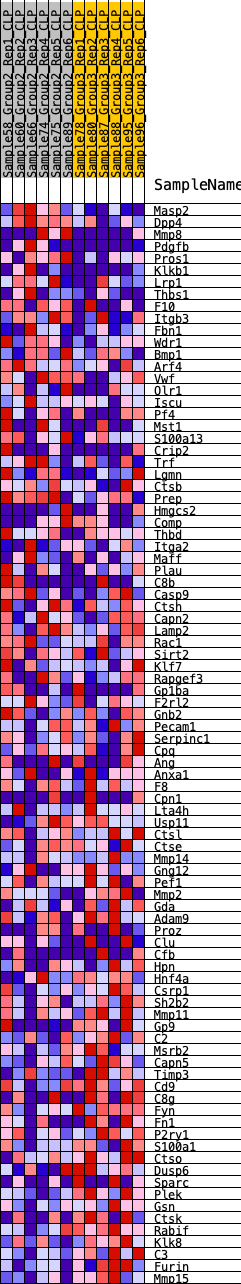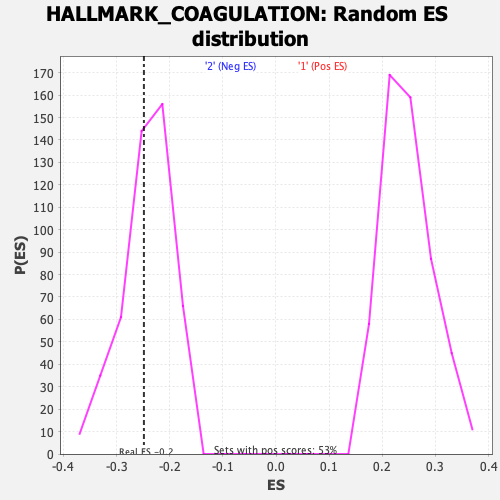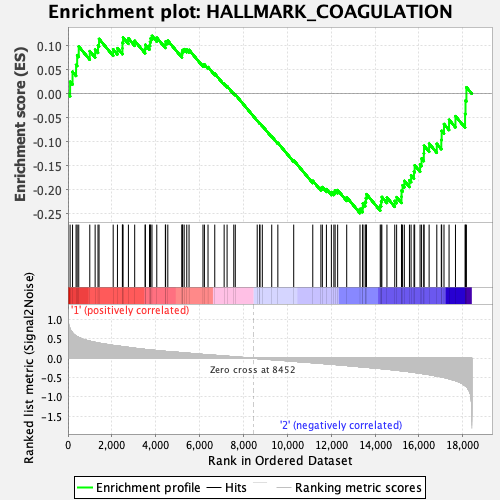
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | CLP.Basophil_Pheno.cls #Group2_versus_Group3.Basophil_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | Basophil_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.24849452 |
| Normalized Enrichment Score (NES) | -1.0283343 |
| Nominal p-value | 0.40552017 |
| FDR q-value | 0.86358136 |
| FWER p-Value | 0.987 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Masp2 | 96 | 0.747 | 0.0251 | No |
| 2 | Dpp4 | 207 | 0.656 | 0.0457 | No |
| 3 | Mmp8 | 368 | 0.572 | 0.0602 | No |
| 4 | Pdgfb | 418 | 0.561 | 0.0803 | No |
| 5 | Pros1 | 490 | 0.537 | 0.0982 | No |
| 6 | Klkb1 | 993 | 0.440 | 0.0887 | No |
| 7 | Lrp1 | 1237 | 0.409 | 0.0920 | No |
| 8 | Thbs1 | 1368 | 0.392 | 0.1009 | No |
| 9 | F10 | 1415 | 0.388 | 0.1141 | No |
| 10 | Itgb3 | 2060 | 0.330 | 0.0924 | No |
| 11 | Fbn1 | 2254 | 0.317 | 0.0947 | No |
| 12 | Wdr1 | 2480 | 0.299 | 0.0945 | No |
| 13 | Bmp1 | 2483 | 0.299 | 0.1066 | No |
| 14 | Arf4 | 2510 | 0.297 | 0.1172 | No |
| 15 | Vwf | 2754 | 0.279 | 0.1152 | No |
| 16 | Olr1 | 3040 | 0.257 | 0.1101 | No |
| 17 | Iscu | 3515 | 0.227 | 0.0934 | No |
| 18 | Pf4 | 3527 | 0.226 | 0.1020 | No |
| 19 | Mst1 | 3715 | 0.215 | 0.1006 | No |
| 20 | S100a13 | 3741 | 0.214 | 0.1079 | No |
| 21 | Crip2 | 3767 | 0.213 | 0.1152 | No |
| 22 | Trf | 3828 | 0.210 | 0.1204 | No |
| 23 | Lgmn | 4047 | 0.197 | 0.1165 | No |
| 24 | Ctsb | 4438 | 0.176 | 0.1024 | No |
| 25 | Prep | 4450 | 0.176 | 0.1089 | No |
| 26 | Hmgcs2 | 4550 | 0.171 | 0.1104 | No |
| 27 | Comp | 5195 | 0.142 | 0.0811 | No |
| 28 | Thbd | 5200 | 0.142 | 0.0866 | No |
| 29 | Itga2 | 5226 | 0.140 | 0.0909 | No |
| 30 | Maff | 5296 | 0.137 | 0.0927 | No |
| 31 | Plau | 5414 | 0.131 | 0.0917 | No |
| 32 | C8b | 5518 | 0.127 | 0.0912 | No |
| 33 | Casp9 | 6153 | 0.097 | 0.0605 | No |
| 34 | Ctsh | 6219 | 0.095 | 0.0608 | No |
| 35 | Capn2 | 6383 | 0.088 | 0.0555 | No |
| 36 | Lamp2 | 6690 | 0.075 | 0.0418 | No |
| 37 | Rac1 | 7118 | 0.057 | 0.0208 | No |
| 38 | Sirt2 | 7252 | 0.052 | 0.0157 | No |
| 39 | Klf7 | 7557 | 0.038 | 0.0006 | No |
| 40 | Rapgef3 | 7626 | 0.034 | -0.0017 | No |
| 41 | Gp1ba | 8625 | -0.007 | -0.0559 | No |
| 42 | F2rl2 | 8737 | -0.011 | -0.0615 | No |
| 43 | Gnb2 | 8750 | -0.012 | -0.0617 | No |
| 44 | Pecam1 | 8858 | -0.016 | -0.0669 | No |
| 45 | Serpinc1 | 9287 | -0.034 | -0.0889 | No |
| 46 | Cpq | 9566 | -0.045 | -0.1022 | No |
| 47 | Ang | 10289 | -0.077 | -0.1385 | No |
| 48 | Anxa1 | 11158 | -0.114 | -0.1813 | No |
| 49 | F8 | 11528 | -0.131 | -0.1961 | No |
| 50 | Cpn1 | 11595 | -0.134 | -0.1943 | No |
| 51 | Lta4h | 11780 | -0.142 | -0.1985 | No |
| 52 | Usp11 | 12006 | -0.152 | -0.2047 | No |
| 53 | Ctsl | 12120 | -0.157 | -0.2045 | No |
| 54 | Ctse | 12177 | -0.160 | -0.2010 | No |
| 55 | Mmp14 | 12292 | -0.165 | -0.2005 | No |
| 56 | Gng12 | 12705 | -0.186 | -0.2155 | No |
| 57 | Pef1 | 13311 | -0.217 | -0.2397 | Yes |
| 58 | Mmp2 | 13435 | -0.224 | -0.2373 | Yes |
| 59 | Gda | 13441 | -0.224 | -0.2284 | Yes |
| 60 | Adam9 | 13547 | -0.230 | -0.2248 | Yes |
| 61 | Proz | 13581 | -0.232 | -0.2172 | Yes |
| 62 | Clu | 13605 | -0.232 | -0.2090 | Yes |
| 63 | Cfb | 14234 | -0.264 | -0.2326 | Yes |
| 64 | Hpn | 14274 | -0.266 | -0.2239 | Yes |
| 65 | Hnf4a | 14307 | -0.268 | -0.2148 | Yes |
| 66 | Csrp1 | 14539 | -0.282 | -0.2160 | Yes |
| 67 | Sh2b2 | 14895 | -0.302 | -0.2231 | Yes |
| 68 | Mmp11 | 14981 | -0.308 | -0.2152 | Yes |
| 69 | Gp9 | 15202 | -0.322 | -0.2142 | Yes |
| 70 | C2 | 15206 | -0.322 | -0.2013 | Yes |
| 71 | Msrb2 | 15252 | -0.325 | -0.1905 | Yes |
| 72 | Capn5 | 15332 | -0.331 | -0.1814 | Yes |
| 73 | Timp3 | 15565 | -0.346 | -0.1800 | Yes |
| 74 | Cd9 | 15640 | -0.351 | -0.1698 | Yes |
| 75 | C8g | 15772 | -0.362 | -0.1622 | Yes |
| 76 | Fyn | 15803 | -0.365 | -0.1491 | Yes |
| 77 | Fn1 | 16050 | -0.383 | -0.1469 | Yes |
| 78 | P2ry1 | 16118 | -0.391 | -0.1347 | Yes |
| 79 | S100a1 | 16221 | -0.399 | -0.1241 | Yes |
| 80 | Ctso | 16229 | -0.400 | -0.1082 | Yes |
| 81 | Dusp6 | 16462 | -0.420 | -0.1038 | Yes |
| 82 | Sparc | 16812 | -0.458 | -0.1043 | Yes |
| 83 | Plek | 17017 | -0.478 | -0.0960 | Yes |
| 84 | Gsn | 17034 | -0.481 | -0.0773 | Yes |
| 85 | Ctsk | 17140 | -0.494 | -0.0630 | Yes |
| 86 | Rabif | 17371 | -0.527 | -0.0541 | Yes |
| 87 | Klk8 | 17663 | -0.575 | -0.0467 | Yes |
| 88 | C3 | 18101 | -0.705 | -0.0419 | Yes |
| 89 | Furin | 18121 | -0.711 | -0.0141 | Yes |
| 90 | Mmp15 | 18162 | -0.731 | 0.0134 | Yes |